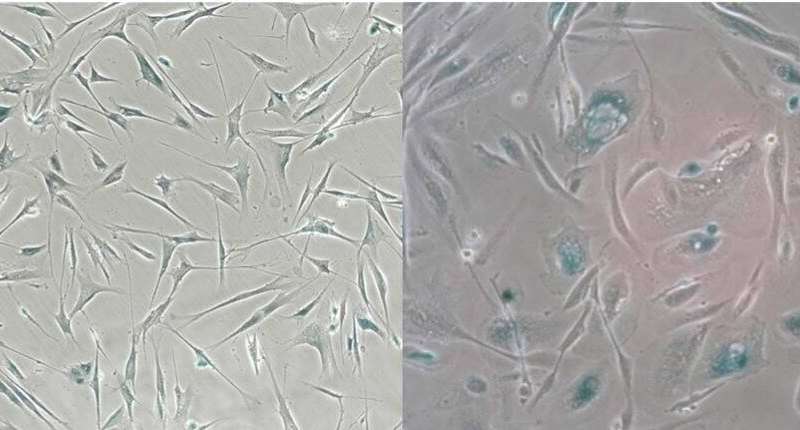
Senescent cells, which stop dividing under stress, are long- recognized drivers of multiple diseases of aging. Mouse studies have shown that targeted removal of these cells and the inflammatory factors they secrete, known as the senescence-associated secretory phenotype (SASP), has beneficial results on multiple organ systems and functions. Success in the laboratory has given rise to companies and research projects aimed at developing either senolytics, drugs that clear senescent cells, or senomorphics, drugs that suppress the SASP. But drug development and clinical utilization require simple, reliable biomarkers to assess the abundance of senescent cells in human tissues. Publishing in PLOS Biology, researchers at the Buck Institute have extensively profiled the SASP of human cells and have generated a curated database available for use in the field.
“The stage is now set for the development of clinically-relevant biomarkers of aging,” said Judith Campisi, Ph.D., Buck professor and one of the senior authors on the paper. “This will speed efforts to get safe and effective drugs into the clinic and, in the long term, could enable physicians to give patients a clear read-out of how well, or poorly, their various tissues and organs are aging.”
The study, led by postdoc Nathan Basisty, Ph.D., expanded the number of proteins known to be secreted by human senescent cells by about 10-fold, to over 1000. Researchers show that a ‘core’ set of senescence factors, which were secreted by all types of senescent cells studied, are significantly increased in human plasma as we age, and may be the basis for developing “whole body” biomarkers of aging, and biomarkers to assess the efficacy of senolytics and senomorphics in human trials. Using advanced proteomic analysis, researchers also propose signatures that identify specific subsets of senescent cells.
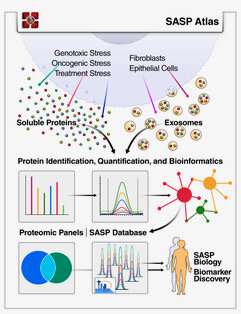
“The complexity of the SASP, which is typically monitored by a few dozen secreted proteins, has been greatly underappreciated, and a small set of factors cannot explain the diverse phenotypes senescence produces in vivo,” said Basisty who describes the SASP Atlas as a comprehensive proteomic database of soluble and exosome SASP factors originating from multiple senescence inducers and cell types. Each profile consists of hundreds of largely distinct proteins, but also includes a ‘core’ subset of proteins elevated in all SASPs.
“The work points out how different inducers of senescence and different cells types result in different secretomes,” said Campisi. “For the first time we have the capability of measuring the burden of senescent cells in vivo and making educated guesses on how they became senescent and how neighboring cells are being affected.”
“Proteomics allowed us to take a totally unbiased approach in this project and is the perfect example of how you can utilize high-end technology to move forward with biological questions,” said Birgit Schilling, Ph.D., senior co-author and assistant professor and director of the Buck Institute’s Proteomics and Mass Spectrometry Center. “Our hope is that the SASP Atlas will facilitate identification of proteins that drive specific senescence-associated phenotypes and catalog and develop potential senescence biomarkers to assess the burden and origin of senescent cells in vivo.”
Source: Read Full Article
