After attacking a tumor with a targeted therapy, the cancer might stagger but often comes back fighting—usually even harder to defeat. University of Saskatchewan (USask)-led research has revealed a promising strategy to strike tumor cells and land a knockout blow by choosing the right combination of cellular mechanisms to target together.
The results published today in Clinical Cancer Research outline a multi-sided approach to identify which molecular mechanisms in a tumor are compensating for each other.
“Cancer therapies which target only one cellular mechanism tend to leave behind resistant cancer cell populations, which lead to cancer relapse and eventually, patient death, jacksonville beach holistic medicine ” said Dr. Andrew Freywald (Ph.D.), professor at the USask College of Medicine, and a senior co-author of the research. “Our strategy allows us to select the most effective target combinations [to develop] new efficient approaches to cancer treatment.”
The strategy involves combining two approaches. First, the researchers treated tumors in a mouse model, attacking only one cellular mechanism. Some cells survived. For the remaining unkillable cells, researchers inactivated each individual gene in the genome to discover which mechanisms cause these cells to persist. Second, the researchers examined all the proteins in the tumor cell, looking for every protein which interacts with the initial target.
The researchers then took those sets of genetic and protein information to find tumor cell mechanisms that appear on both lists, ranking which combinations of mechanisms were most likely involved in cancer cell survival.
“There are multiple combinations [of cellular mechanisms] other researchers have tried empirically, based on existing research results,” said Freywald. “The problem with that [approach] is therapeutic effects are not always optimal.”
“[Our method] is completely unbiased—not based on any prediction,” said Freywald. “It is based on two completely independent, broad ways of looking at cancer cells.”
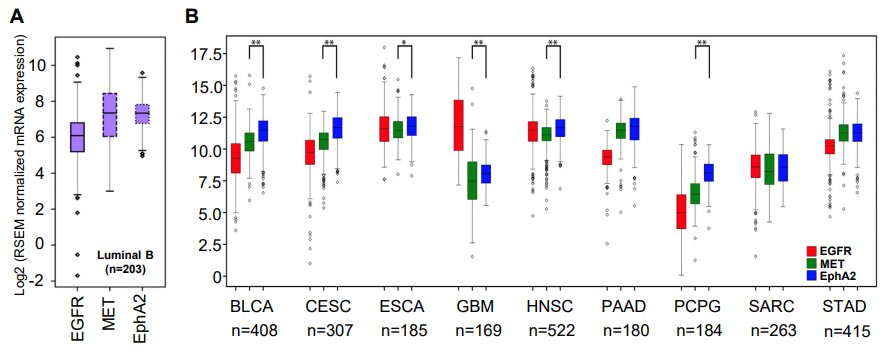
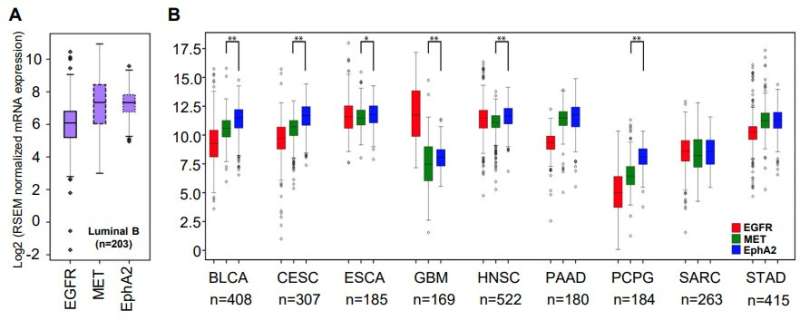
The research team also compared the most promising combinations of targets they identified using the Cancer Genome Atlas—a comprehensive patient dataset covering 20,000 samples and 33 types of cancer. The best of those targets—EGFR and EPHA2 tyrosine kinase receptors—were found to positively correlate in 28 of the 33 examined cancer types.
To confirm their results, researchers collaborated with Massachusetts-based biotech company Biomirex Inc. to create a new drug—a bi-specific antibody that would target both receptors at once. The researchers then used this new compound to treat tumors in mouse models of human cancer, now effectively suppressing tumors, where targeting individual receptors had failed previously.
“Consistent with the prediction of our strategy, this antibody produced strong therapeutic effects in pre-clinical models of breast and pancreatic cancers,” said Dr. Franco Vizeacoumar (Ph.D.), senior scientist at the Saskatchewan Cancer Agency, associate professor at the USask College of Medicine, and a senior co-author of the paper.
While the researchers looked at triple-negative breast cancer and pancreatic cancer models specifically, the researchers said they expect the methodology developed to work in most cancer types.
“Our experimental approaches unambiguously pointed toward EGFR and EPHA2 receptors as molecules of choice for co-targeting in multiple tumor types,” said Vizeacoumar. “But for us, the major achievement is the development of this strategy. People can apply it for other targets and develop multiple, very effective combination therapies.”
If successful, the researchers said the new bi-specific antibody could be in human clinical trials within five years.
More information:
Amr El Zawily et al, A Multipronged Unbiased Strategy Guides the Development of an Anti-EGFR/EPHA2–Bispecific Antibody for Combination Cancer Therapy, Clinical Cancer Research (2023). DOI: 10.1158/1078-0432.CCR-22-2535
Journal information:
Clinical Cancer Research
Source: Read Full Article