Researchers in the United States have demonstrated that the viral spike protein used by severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) to infect host cells adopts an open conformation exposing novel epitopes that could serve as targets for therapeutic strategies to protect against coronavirus disease 2019 (COVID-19).
The currently approved COVID-19 vaccines and many clinical diagnostic techniques are based on the SARS-CoV-2 spike ectodomain that stabilizes the protein in the prefusion conformation.
Now, a team from the University of California in Berkeley, the University of Texas at Austin, and Chan Zuckerberg Biohub in San Francisco have identified a new conformation exposing the conserved trimer interface that is otherwise buried in the prefusion state.
Subsequently, this exposes potential epitopes for pan-coronavirus antibody and ligand recognition, says Susan Marqusee and colleagues.
“Knowledge of the structure and populations of this conformation will help improve existing diagnostics, therapeutics, and vaccines,” they write.
A pre-print version of the research paper is available on the bioRxiv* server, while the article undergoes peer review.
More about the viral spike protein
The SARS-CoV-2 spike is a large trimeric protein that mediates the initial stage of the infection process when its receptor-binding domain (RBD) attaches to the host cell receptor angiotensin-converting enzyme 2 (ACE2).
Shortly following the release of the SARS-CoV-2 genome sequence in January 2020, a version of the spike ectodomain – S-2P – was developed that stabilizes the prefusion conformation.
This S-2P ectodomain has been widely used for vaccine development and in many studies of the structure and function of the spike ectodomain. More than 250 structures of SARS-CoV2 spike ectodomains are now available in the Protein Data Bank.
“These structural studies together with other functional studies demonstrate that, like all class 1 viral fusion proteins, the spike protein is dynamic, sampling several different conformations during its functional lifecycle,” says Marqusee and colleagues.
Three individual RBDs adopt an “up” and a “down” conformation, with the up conformation exposing the ACE2-binding motif required for infectivity.
Following receptor binding and cleavage between subunits S1 and S2, the spike undergoes a re-folding event to enable fusion, before then adopting a stable post-fusion conformation.
However, Marqusee and colleagues say there are very few experimental studies providing information on the dynamics of the prefusion state.
“Experimentally, the conformational landscape of spike has not been well interrogated and the effects of perturbations, such as ligand binding (both receptor and antibodies) or amino acid substitutions found in emerging variants of concern are unknown,” writes the team.
What did the researchers do?
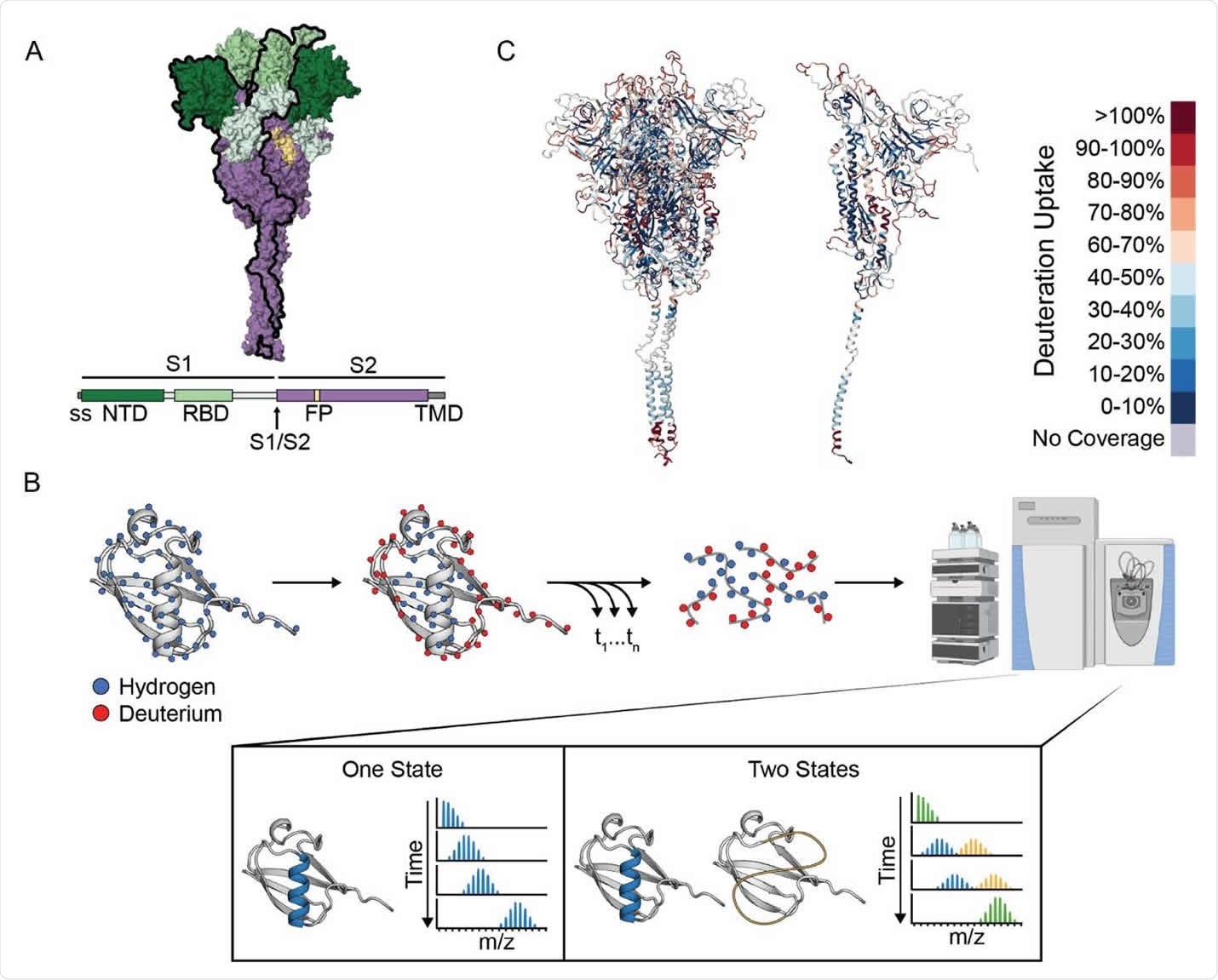
The researchers used hydrogen deuterium exchange monitored by mass spectrometry (HDX-MS) to investigate the landscape of the spike prefusion ectodomain and the effects of ligand binding and sequence variation.
“HDX-MS offers an ideal complement to the ever-growing number of structural studies on the SARS-CoV-2 spike protein, providing information on its conformational ensemble and dynamics,” says the team.
The results indicated that the spike protein populates two conformations within the prefusion state – the canonical prefusion structure (state A) and another unexpected conformation with a more exposed open-trimer interface (state B).
When the researchers used temperature to perturb the structure, the two conformations were found to interconvert reversibly, with a preference for state B at 4 °C and for state A at 37 °C.
The new conformation is an open trimer
State B exposes the conserved S2 trimer interface that is otherwise buried in the prefusion conformation, thereby providing new potential epitopes for pan-coronavirus antibody and ligand recognition.
To test whether mutations can influence the interconversion of the A and B conformation, the team monitored the conversion for a variant of HexaPro that includes five mutations found in S1 of the B.1.1.7 (alpha) variant that emerged in the UK.
Indeed, the UK S1 HexaPro exhibited differences in both the relative preference for state B and the kinetics of interconversion.
At 4 °C, UK S1 HexaPro converted to state B almost 20 times more rapidly than the original HexaPro.
“Thus, despite their location in the S1 subunit and not at the core trimer interface, these specific B.1.1.7 mutations allosterically affect the interconversion of these two states,” write the researchers.
Testing the effects of a recently developed antibody
A recently developed antibody called 3A3 that binds to both SARS-CoV-1 and SARS-CoV-2 targets an epitope present in a region of the spike (around residues 980 to 1000) for which Marqusee and colleagues had previously observed bimodal behavior.
However, this region had been considered inaccessible since it is buried in the prefusion structure when all RBDs are down and is also occluded when the RBDs are up.
Interestingly, the HDX data indicated that this region is exposed after conversion to state B.
When the team repeated the HDX experiment in the presence of 3A3, a significantly increased level of protection was observed in the 978–1001 region that was directly associated with state B.
What did the authors conclude?
The researchers say that further understanding of this newly identified state B will help to improve the development of diagnostics, therapeutics, and vaccines.
“The data collectively suggest that state B exposes broadly neutralization-sensitive epitopes that may be of interest for future therapeutics and vaccines,” says Marqusee and colleagues.
Moreover, these epitopes are found in the most highly conserved part of the spike – the S2 trimer interface – and therefore present an ideal target for vaccines that would provide protection across different coronaviruses.
*Important Notice
bioRxiv publishes preliminary scientific reports that are not peer-reviewed and, therefore, should not be regarded as conclusive, guide clinical practice/health-related behavior, or treated as established information.
- Marqusee S, et al. The SARS-CoV-2 spike reversibly samples an open-trimer conformation exposing novel epitopes. bioRxiv, 2021. doi: https://doi.org/10.1101/2021.07.11.451855, https://www.biorxiv.org/content/10.1101/2021.07.11.451855v1
Posted in: Medical Research News | Disease/Infection News
Tags: ACE2, Amino Acid, Angiotensin, Angiotensin-Converting Enzyme 2, Antibodies, Antibody, Cell, Coronavirus, Diagnostic, Diagnostics, Enzyme, Genome, Ligand, Mass Spectrometry, Protein, Receptor, Research, Respiratory, SARS, SARS-CoV-2, Severe Acute Respiratory, Severe Acute Respiratory Syndrome, Spectrometry, Spike Protein, Syndrome, Therapeutics, Vaccine

Written by
Sally Robertson
Sally first developed an interest in medical communications when she took on the role of Journal Development Editor for BioMed Central (BMC), after having graduated with a degree in biomedical science from Greenwich University.
Source: Read Full Article
