In a recent study published in the journal Cell, researchers demonstrated that antibodies generated due to breakthrough infection (BTI) by severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) new variant of concern (VOC) Omicron in vaccinated individuals could neutralize all VOCs.
 Study: Potent cross-reactive antibodies following Omicron breakthrough in vaccinees. Image Credit: Naeblys / Shutterstock
Study: Potent cross-reactive antibodies following Omicron breakthrough in vaccinees. Image Credit: Naeblys / Shutterstock
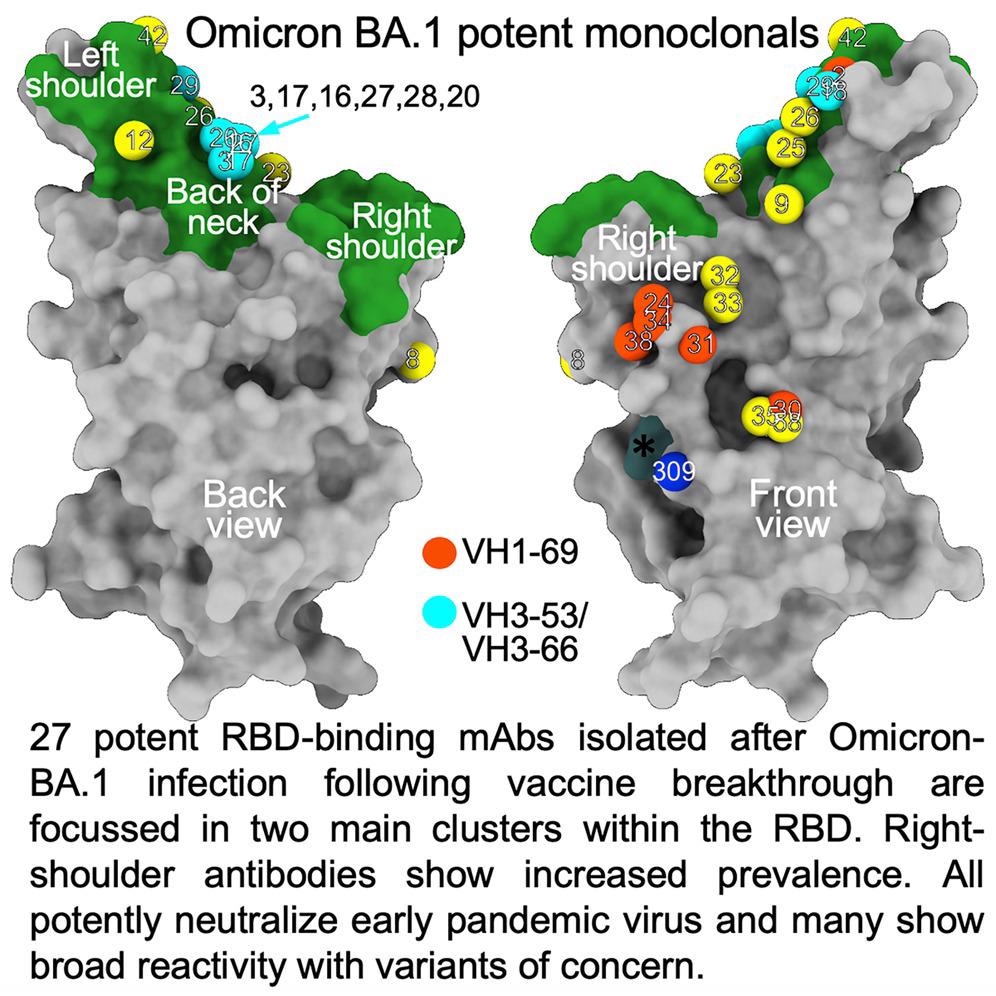
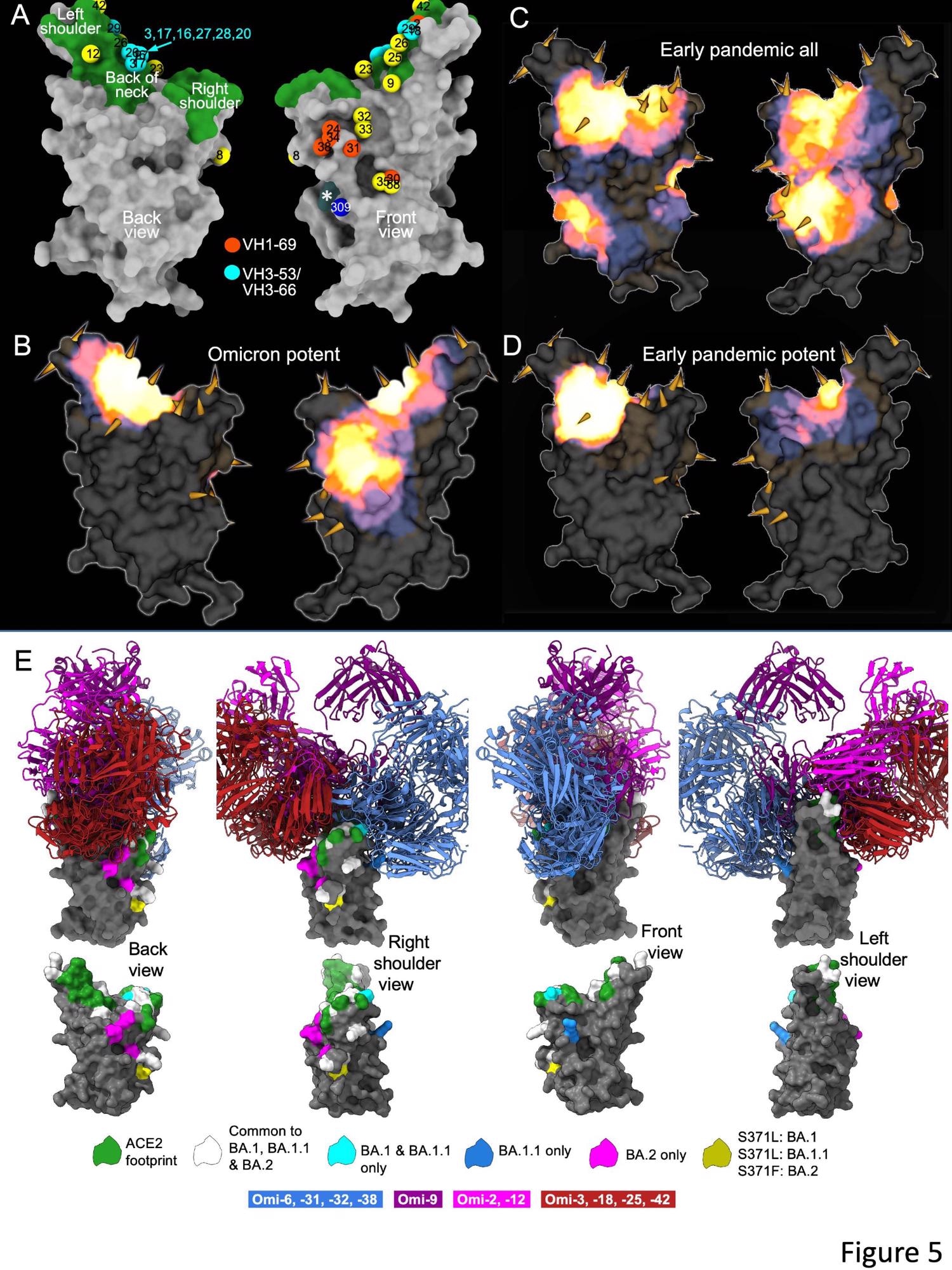
The researchers isolated 27 cross-reactive and broadly potent monoclonal antibodies (mAbs) from vaccinated individuals who contracted an Omicron BTI. Upon structural and functional analysis, these mAbs were located in two clusters within the receptor-binding domain (RBD) of the SARS-CoV-2 spike (S) protein.
Background
The RBD mutations prevalent in all VOCs primarily serve two functions. First, they enhance affinity to angiotensin-converting enzyme 2 (ACE2) receptors on the host cell surface, which increases transmissibility, observed for SARS-CoV-2 Alpha, Beta, and Gamma variants. Second, the RBD mutations confer evasion capability from neutralizing antibodies (nAbs) generated in response to vaccination or previous SARS-CoV-2 infection, observed for Omicron.
Currently, three Omicron sub-variants, BA.1, BA.1.1, and BA.2, are circulating globally. Omicron BA.1.1 has an additional R346K mutation, while BA.2 has six unique mutations in the RBD and is now becoming dominant in several countries. The mutational burden in Omicron is exceptionally concerning as it disrupts the neutralizing capacity of serum from natural infection or vaccination to a much greater extent.
Fortunately, a three-dose coronavirus disease 2019 (COVID-19) vaccination regimen generates reasonable anti-Omicron neutralizing antibodies (nAbs) titers, decreasing the probability of hospitalization and severe disease.
About the study
In the present study, researchers collected sera from 41 vaccinees who received the AZD1222 COVID-19 vaccine and 20 vaccinees who received the messenger ribonucleic acid (mRNA)-based BNT162b2 vaccine post-28 days of receiving the third dose.
They performed neutralization assays on Victoria, an early SARS-CoV-2 isolate containing an S247R substitution in the N-terminal domain (NTD), and BA.1, BA.1.1, and BA.2 Omicron sub-variants. Next, the team collected serum from individuals infected with Omicron BA.1 at two-time points to assess the neutralization profile across all VOCs.
They collected 12 and 16 samples 14 and 21 days from symptom onset, respectively. Of these individuals, all had received a minimum of two vaccine doses, and three had received the third dose following Omicron BTI.
The researchers generated a panel of human mAbs from volunteers who had received two doses of the BNT162b2 vaccine and then contracted a BTI from Omicron BA.1. Further, they stained B cells from five donors with a full-length BA.1 S trimer to sort single cells by fluorescence-activated cell sorting (FACS). The team assembled heavy and light chain sequences into expression vectors using the Gibson reaction. Next, they transfected these vectors into 293T cells after a degenerate reverse transcription-polymerase chain reaction (RT-PCR) to screen culture supernatants for reactivity to BA.1 S or the S of wild type (wt) Wuhan strain together with BA.1 RBD and NTD. They sorted 1,122 single cells and recovered 545 mAbs.
Lastly, using the focus reduction neutralization test (FRNT), the researchers selected the 28 most potent antibodies, with the serum dilution required to reduce virus foci by 50% (FRNT50) titers less than 100 ng/ml for Omicron BA.1.
Study findings
The recipients of the AZD1222 vaccine showed a small but significant reduction in nAb titers against BA.1.1 and BA.2 relative to BA.1. The fold-reduction for BA.1 vs. BA.1.1 was 1.5 after the AZD1222 and BNT162b2 vaccination. Likewise, the fold-reduction for BA.1 vs. BA.2 was 1.4 and 1.2 after the AZD1222 and BNT162b2 vaccination, respectively.
Expectedly, at 14 days from symptom onset, all vaccinated individuals had sera to broadly neutralize all VOCs, except Omicron, with FRNT50 values under 1/1000. At 21 days after symptom onset, titers increased against all variants, including 3.1-fold for BA.1, indicating that Omicron BTI elicited broad-spectrum nAbs.
An enzyme-linked immunosorbent assay (ELISA) showed cross-reactivity between wt and BA.1 S of almost all mAbs. Moreover, a higher proportion of mAbs was RBD-reactive, and the remaining 50% of antibodies, 129 of 545, bound the NTD. ELISA also showed that except for Omi-30 and Omi-41, all mAbs reduced the interaction of RBD with ACE2. Nine out of 28 monoclonals (30%) belonged to the IGHV3-53 and related IGHV3-66 gene families, which typically bind a site at the back of the neck of the RBD and block ACE2 binding.
The Omicron mAb set had around one-half of the gene families observed in the potent early pandemic antibodies. Although IGHV1-69 did not feature in the set of potent early antibodies, there were six IGHV1-69 antibodies in 27 potent RBD binding mAbs. The authors also noted higher levels of somatic mutation in both heavy and light chains of Omicron mAbs relative to the early pandemic set of antibodies.
The most potent of the 28 mAbs, Omi-12, belonged to the IGHV1-58 gene family, isolated from SARS-CoV-2-infected individuals. It cross-neutralized all VOCs while other IGHV1-58 antibodies lost activity against BA.1; however, somatic mutation recovered this potency.
Conclusions
The proportion of Omicron infections caused by BA.2 is surging in several countries, but there is no clinical evidence of increased disease severity. Since there have been many deaths due to Omicron BTIs, it has put considerable pressure on public healthcare systems.
The differences in the S antigenicity of the Omicron sub-lineages might have driven the transmission or increased BA.2 receptor affinity. Compared to BA.1, there was an increase in BA.2 RBD affinity for ACE2 and a reduction in neutralization titers of BA.2 in vaccine serum. However, intriguingly, BTIs due to Omicron in previously vaccinated individuals led to a broad antibody response against all VOCs, including Omicron lineages. Nevertheless, all 27 mAbs showed broad reactivity against all VOCs, most likely due to vaccine memory responses.
Overall, the structural analysis of the current study demonstrated that the RBD has space for the binding of potent mAbs that could broadly neutralize VOCs. It also illustrated the flexibility of the public antibody responses through IGHV3-53/66 and IGHV1-58 gene families. Interestingly, the somatic mutation restored the neutralizing activity against BA.1 and other VOCs.
- Nutalai, R., Zhou, D., Tuekprakhon, A., Ginn, H.M., Supasa, P., Liu, C., Huo, J., Mentzer, A.J., Duyvesteyn, H.M.E., Dijokaite-Guraliuc, A., Skelly, D., Ritter, T.G., Amini, A., Bibi, S., Adele, S., Johnson, S.A., Constantinides, B., Webster, H., Temperton, N., Klenerman, P., Barnes, E., Dunachie, S.J., Crook, D., Pollard, A.J, Lambe, T., Goulder, P., OPTIC consortium, ISARIC4C consortium, Paterson, N.G., Williams, M.A., Hall, D.R., Mongkolsapaya, J., Fry, E.E., Dejnirattisai, W., Ren, J., Stuart, D.I., Screaton, G.R, Potent cross-reactive antibodies following Omicron breakthrough in vaccinees, Cell (2022), DOI: https://doi.org/10.1016/j.cell.2022.05.014, https://www.cell.com/cell/fulltext/S0092-8674(22)00598-0
Posted in: Medical Research News | Disease/Infection News
Tags: ACE2, Angiotensin, Angiotensin-Converting Enzyme 2, Antibodies, Antibody, Assay, Cell, Cell Sorting, Coronavirus, Coronavirus Disease COVID-19, covid-19, Enzyme, Fluorescence, Gene, Healthcare, Mutation, Neck, Omicron, Pandemic, Polymerase, Polymerase Chain Reaction, Protein, Receptor, Respiratory, Ribonucleic Acid, SARS, SARS-CoV-2, Severe Acute Respiratory, Severe Acute Respiratory Syndrome, Syndrome, Transcription, Vaccine, Virus

Written by
Neha Mathur
Neha is a digital marketing professional based in Gurugram, India. She has a Master’s degree from the University of Rajasthan with a specialization in Biotechnology in 2008. She has experience in pre-clinical research as part of her research project in The Department of Toxicology at the prestigious Central Drug Research Institute (CDRI), Lucknow, India. She also holds a certification in C++ programming.
Source: Read Full Article
