Researchers from Ceres Nanosciences, USA, have presented a nanotrap enrichment workflow for capturing and concentrating severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) particles from diagnostic and contrived samples.
This approach enhances both column and magnetic bead-based RNA extraction protocols, providing significant improvements in sequencing results on the Oxford Nanopore technologies (ONT) MinION sequencing platform. Sequencing output demonstrated significantly more mapped viral reads, and 20-80% improved sequence coverage from diagnostic samples with low virus titers.
 Study: Nanotrap Particles Improve Nanopore Sequencing of SARS-CoV-2 and Other Respiratory Viruses. Image Credit: Unitone Vector/Shutterstock
Study: Nanotrap Particles Improve Nanopore Sequencing of SARS-CoV-2 and Other Respiratory Viruses. Image Credit: Unitone Vector/Shutterstock
The team also demonstrated the same approach to enrich two other respiratory viruses namely Influenza A virus and respiratory syncytial virus, highlighting its potential to improve real-time PCR and sequencing results for respiratory viruses other than SARS-CoV-2.
Background
Continuous genetic evolution of SARS-CoV-2 highlights the need to constantly identify and monitor the emergence of new viral variants, and thus specifically demands for rapidly deployable accurate sequencing technologies.
Here, the ONT MinION sequencer comes into focus. Being a portable and relatively inexpensive detection strategy, it can be used for rapid on-site detection and characterization of viral variants in the field. However, the usefulness of these portable sequencing tools is limited by the base-calling accuracy issues that arise in samples with insufficient nucleic acid concentration.
The team from Ceres Nanosciences, in their recent research published on the preprint server bioRxiv*, has addressed this limitation by using an affinity-based magnetic hydrogel particle enrichment technology (Nanotrap particle enrichment technology) that can concentrate viral particles, leading to improved detection of many viral agents including SARS-CoV-2. With superior base-calling, bioinformatic tools can confidently distinguish the genetic mutation in the variant from machine error.
What did the researchers do?
The team developed three workflows to demonstrate the robustness and the ease-of-use of Nanotrap particle technology: a manual method with a column-based RNA extraction (Nanotrap Particle Workflow 1), a manual method with a magnetic-bead-based RNA extraction (Nanotrap Particle Workflow 2), and an automated method with a magnetic-bead-based RNA extraction (Nanotrap Particle Workflow 3).
These workflows were carried out in SARS-CoV-2 contrived (spiked with 1:10 serial dilutions of heat-inactivated SARS-CoV-2 starting from 106 TCID50/mL to 102 TCID50/mL) viral transport media (VTM) as well as remnant diagnostic VTM samples. The Nanotrap particle workflows (+NT) were compared against the workflows without any Nanotrap particles (-NT).
Extracted RNA was prepared for sequencing using the ARTIC nCoV-2019 sequencing protocol and sequenced on the ONT Mk1C sequencing platform, followed by data analysis.
Additionally, real-time RT-PCR was performed on RNA extracted from all three workflows to identify a potential correlation between the two assays and confirm the presence of SARS-CoV-2.
Nanotrap particles improve sequencing results
With Nanotrap Workflow 1, Nanotrap particles significantly improved sequencing results at various virus concentrations when compared to the workflow without Nanotrap particles. A 6.0X increase in reads mapped to SARS-CoV-2 was observed at 106 TCID50/mL and a 2.0X increase was seen at 105 TCID50/mL. These improvements were statistically significant. Viral detection in real-time qPCR was also significantly improved by two PCR Cycle thresholds (Ct) across the first four serial dilutions.
As column-based RNA extractions are more appropriate when working with less number of samples, the team tried to assess the Nanotrap particles’ ability to improve on sequencing when using RNA extracted via magnetic beads (Nanotrap Workflow 2). Here, an improvement of 1.9X in SARS-CoV-2 mapped reads was observed at 106 TCID50/mL and an improvement of 1.4X at 105 TCID50/mL. Additionally, real-time qPCR detection of SARS-CoV-2 was enhanced, providing an average 1.5 Ct improvement at 106-102 TCID50/mL.
“RT-PCR results showed Nanotrap particle enrichment was efficacious for lower concentration samples for both workflows, potentially suggesting that on an alternative sequencing platform with greater overall sensitivity, Nanotrap particles could also improve sequencing of these lower titer virus samples”, the team highlights.
To test the efficacy of Nanotrap particles with real clinical samples, the team evaluated the Nanotrap particle workflows using 10 diagnostic remnant clinical swab samples with reported qPCR Ct values ranging from 24 to 35. The team quantified both total viral mapped reads and the associated percent genome coverage at 30x depth of the processed samples.
Nanotrap Particle Workflow 1 resulted in a significant 7X improvement in total viral reads mapped across all 10 diagnostic remnant samples, which further resulted in an average viral genome coverage increase of 52% over samples processed without Nanotrap particles. RT-PCR confirmed the presence of SARS-CoV-2 with an average improvement of 4 Ct over no Nanotrap particle samples.
Automated Nanotrap Particle Workflow 3 method on automated KingFisher Apex System also demonstrated improved results, with an average improvement of 42X in total viral mapped reads that corresponded to an average 51% increase in viral genome coverage relative to (-NT) samples. Again, qPCR confirmed the presence of SARS-CoV-2 for all 10 samples with an average 3.7 Ct improvement.
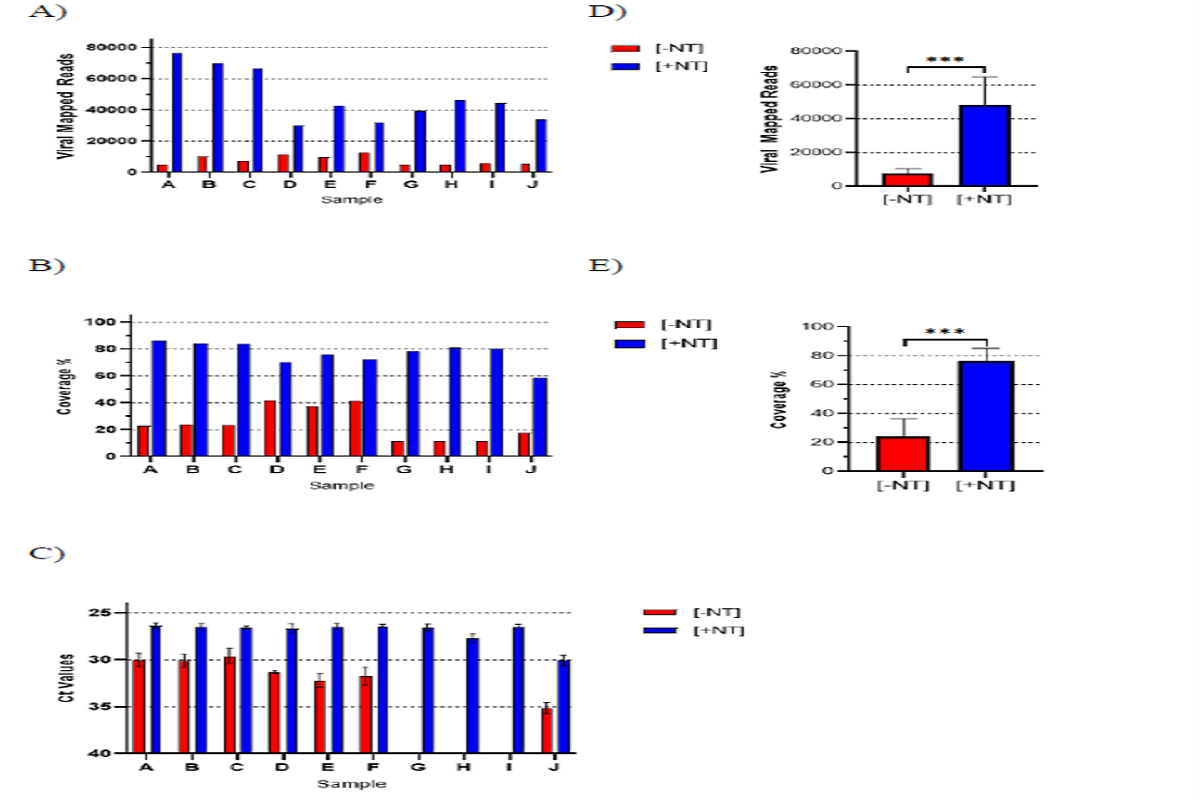 Figure one: Nanotrap Particle Workflow 1 Improves Sequencing of Diagnostic Remnant SARS-CoV-2 Samples. 10 SARS-CoV-2 positive diagnostic remnant samples were processed using Nanotrap Particle Workflow 1 [+NT] or the RNEasy Kit alone [-NT]. Samples then underwent sequencing on a ONT MinION R.9 flow cell and were analyzed by Viral Mapped Reads to SARS-CoV-2 (A), Viral Genome Coverage at 30x depth (B), or RT-PCR (C). [+NT] were compared to [-NT] by paired t-test in order to assess significance of increased viral detection (D),(E). ***p<0.001.
Figure one: Nanotrap Particle Workflow 1 Improves Sequencing of Diagnostic Remnant SARS-CoV-2 Samples. 10 SARS-CoV-2 positive diagnostic remnant samples were processed using Nanotrap Particle Workflow 1 [+NT] or the RNEasy Kit alone [-NT]. Samples then underwent sequencing on a ONT MinION R.9 flow cell and were analyzed by Viral Mapped Reads to SARS-CoV-2 (A), Viral Genome Coverage at 30x depth (B), or RT-PCR (C). [+NT] were compared to [-NT] by paired t-test in order to assess significance of increased viral detection (D),(E). ***p<0.001.
Enhanced detection of other respiratory viruses
To confirm the enriching ability of Nanotrap particles for other viruses and not just SARS-CoV-2, the team similarly tested RNA extraction with workflow 1 on contrived VTM samples spiked with Influenza A (H1N1) virus and RSV. Noticeably, Nanotrap particles were able to improve sequencing results for both viruses. As with SARS-CoV-2, Nanotrap particles increased viral mapped reads by 4X for both Influenza A and RSV compared to the samples processed without Nanotrap particles. Additionally, Nanotrap particles significantly improved the detection of viral RNA with qPCR. Based on this experiment, the team suggests that Nanotrap particles can be used to identify and improve sequencing results of multiple viruses in VTM samples.
Sequencing library preparation protocol involves the use of various enzymes that can be negatively impacted by the presence of detrimental biological material present in the real human-derived diagnostic samples. Pre-processing with Nanotrap particles enables the capture of viral material while reducing host cell debris and other contaminating material. The team thus suggests that workflows without Nanotrap particles are more likely to be impacted by inhibition.
Conclusions
As the viral genome coverage was greatly increased and was almost improved by 80% in the majority of diagnostic samples, the team suggests that using Nanotrap particles can allow for the accommodation of more clinical samples in a single run.
The team highlights the need for a simple and rapid workflow in order to achieve a high sample throughput in a public health setting and therefore recommends the automated Nanotrap Workflow 3 for such settings.
It is worth noting that this automated method would enable the processing of 96 samples in 1 hour, which is significantly faster and far more user friendly than the manual-column extraction method, making this an attractive proposition for medium- to high-throughput laboratories”
Additionally, Nanotrap particle workflows can be used for the improvement of broad-scale viral detection by sequencing as demonstrated from the sequencing experiments conducted on Influenza A and RSV.
Overall, the observations suggest that Nanotrap particle enrichment allows for sequencing of lower titer clinical samples in VTM using the ONT MinION Sequencer, which otherwise may not have been suitable for sequencing.
A Nanotrap particle concentrating method paired with a ONT sequencing platform allows for a powerful sequencing tool that could potentially be deployed in an area with lack of access to more traditional sequencing or sample processing equipment” the team concludes.
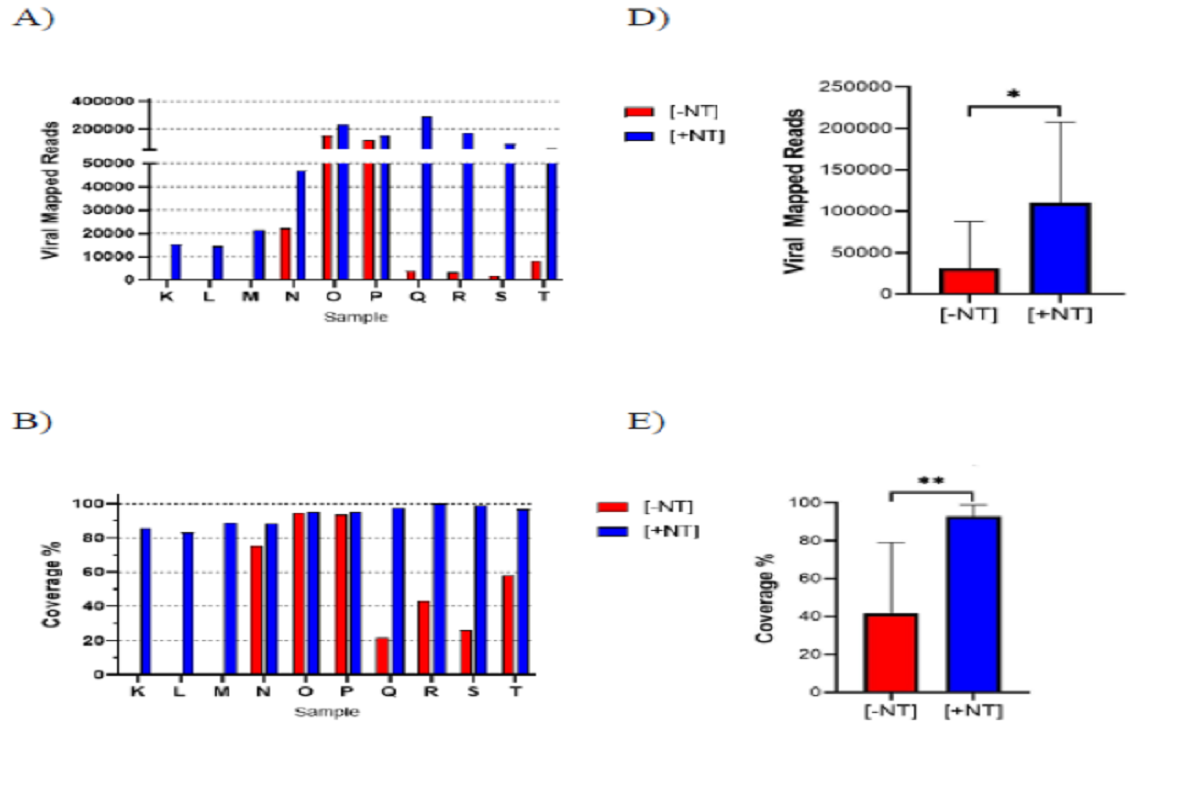 Figure two: Nanotrap Particle Workflow 3 Improves Sequencing Output of Diagnostic Remnant SARS-CoV-2 Samples. 10 SARS-CoV-2 positive diagnostic remnant samples were processed using Nanotrap Particle Workflow 3 [+NT] or the MagMAX kit alone [-NT]. Samples then underwent sequencing on a ONT MinION R.9 flow cell and were analyzed by Viral Mapped Reads to SARS-CoV-2 (A), Viral Genome Coverage at 30x depth (B), or RT-PCR (C). [+NT] were compared to [-NT] by paired t-test in order to assess significance of increased viral detection (D), (E). * p<0.05, ** p<0.01.
Figure two: Nanotrap Particle Workflow 3 Improves Sequencing Output of Diagnostic Remnant SARS-CoV-2 Samples. 10 SARS-CoV-2 positive diagnostic remnant samples were processed using Nanotrap Particle Workflow 3 [+NT] or the MagMAX kit alone [-NT]. Samples then underwent sequencing on a ONT MinION R.9 flow cell and were analyzed by Viral Mapped Reads to SARS-CoV-2 (A), Viral Genome Coverage at 30x depth (B), or RT-PCR (C). [+NT] were compared to [-NT] by paired t-test in order to assess significance of increased viral detection (D), (E). * p<0.05, ** p<0.01.
*Important notice
bioRxiv publishes preliminary scientific reports that are not peer-reviewed and, therefore, should not be regarded as conclusive, guide clinical practice/health-related behavior, or treated as established information.
- Andersen P. et al. (2021). Nanotrap Particles Improve Nanopore Sequencing of SARS-CoV-2 and Other Respiratory Viruses. bioRxiv. doi: https://doi.org/10.1101/2021.12.08.471814, https://www.biorxiv.org/content/10.1101/2021.12.08.471814v1
Posted in: Medical Science News | Medical Research News | Disease/Infection News
Tags: Cell, Coronavirus, Coronavirus Disease COVID-19, CT, Diagnostic, Efficacy, Evolution, Genetic, Genome, H1N1, heat, Hydrogel, Influenza, Library Preparation, Magnetic Beads, Mutation, Nucleic Acid, Public Health, Research, Respiratory, Respiratory Syncytial Virus, RNA, RNA Extraction, SARS, SARS-CoV-2, Severe Acute Respiratory, Severe Acute Respiratory Syndrome, Syndrome, Virus

Written by
Namita Mitra
After earning a bachelor’s degree in Veterinary Sciences and Animal Health (BVSc) in 2013, Namita went on to pursue a Master of Veterinary Microbiology from GADVASU, India. Her Master’s research on the molecular and histopathological diagnosis of avian oncogenic viruses in poultry brought her two national awards. In 2013, she was conferred a doctoral degree in Animal Biotechnology that concluded with her research findings on expression profiling of apoptosis-associated genes in canine mammary tumors. Right after her graduation, Namita worked as Assistant Professor of Animal Biotechnology and taught the courses of Animal Cell Culture, Animal Genetic Engineering, and Molecular Immunology.
Source: Read Full Article
