Autoimmune diseases are conditions in which the immune system mistakenly attacks the body. Scientists know from previous research that the composition of the gut microbiome, the billions of microorganisms living in the human digestive system, is linked to the development of autoimmune diseases. However, the contribution of the gut virome—the viruses living in our gastrointestinal tract—in autoimmune diseases is unknown.
In a study recently published in Annals of the Rheumatic Diseases, researchers from Osaka University Graduate School of Medicine have shown that in people suffering from autoimmune diseases, the composition of the gut virome is compromised.
Autoimmune diseases cause significant chronic morbidity and disability worldwide. They include rheumatoid arthritis, which is most common in older adults, and systemic lupus erythematosus, which is relatively prevalent among young women. The human gastrointestinal tract contains diverse populations of bacteria, fungi, viruses, and other microorganisms collectively called the gut microbiome. It is now widely recognized that the gut microbiome markedly influences our health via the immune and metabolic systems.
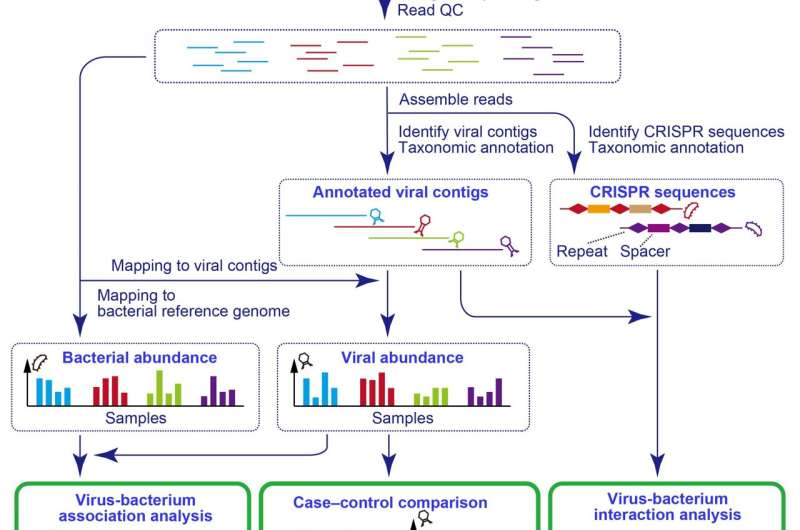
Although they make up a large proportion of the gut microbiome, the contribution of viruses to the effects on health has been far less studied than that of the bacterial component of the gut microbiome because of the technical difficulty of studying these tiny entities. The most predominant component of the gut virome are bacteriophages, viruses that infect bacteria and can alter their physiological function. The Osaka team aimed to investigate the role of bacteriophages in the gut microbiome of individuals with autoimmune diseases in order to reveal a potential link.
The researchers analyzed the gut virome of 476 Japanese individuals, including 111 patients with rheumatoid arthritis, 47 patients with systemic lupus erythematosus, 29 patients with multiple sclerosis, and 289 healthy control volunteers. They constructed a new analytic pipeline to recover viral sequences from whole-metagenome shotgun sequencing data. This enabled them to quantify the abundance of the viruses that reside within the gut environment, providing an invaluable tool to study viruses, which are otherwise difficult or impossible to analyze.
“Our case-control comparison of viral abundance revealed that crAss-like phages, which are one of the main components of a healthy gut virome, were significantly less abundant in the gut of the patients with autoimmune disease, particularly in patients with rheumatoid arthritis and systemic lupus erythematosus,” says Yoshihiko Tomofuji, lead author of the study.
The investigators went on to use CRISPR-based analysis to study possible bacterial targets of crAss-like phages. They observed that the virus called Podoviridae, which has a symbiotic relationship to the bacteria Faecalibacterium, significantly decreased in the gut of the patients with systemic lupus erythematosus. “These analyses have revealed a previously missing part of the autoimmunity-associated gut microbiome and presented new candidates that contribute to the development of autoimmune diseases,” explains Yukinori Okada, senior author of the study.
Source: Read Full Article
