Using simulations, a team of researchers in the U.S. found that the glycan at position N343 of the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) spike protein’s receptor-binding domain (RBD) helps in its opening. A mechanism that aids its binding to the host cell receptor.
The key structure of the SARS-CoV-2 that allows it to infect host cells is the spike protein, which is present on the virus surface. Among the several structures of the spike protein defined, most are conformations where the RBD is in the up or down configuration. The RBD is responsible for interacting with the angiotensin-converting enzyme 2 (ACE2) of the host cells and mediating virus entry.
For viral entry, the RBD transitions from a down state to an up state, making it accessible to ACE2. Previous studies have shown that the spike protein transitions between four different conformations, depending on the presence of ACE2 and antibodies.
Molecular dynamics (MD) simulations indicated the spike protein is shielded by glycans, and glycans at N165 and N234 support the RBD in the open configuration. Although several MD simulations have been performed, they do not include glycosylation of the spike protein or need significant sampling on the order of milliseconds. Cryo-electron microscopy (EM) has revealed the up and down conformations of the RBD, but it is difficult to study the RBD opening process experimentally.
Simulating how spike protein opens
In a new paper published on the bioRxiv* preprint server, researchers report the results of simulations on the RBD opening pathway for the fully glycosylated SARS-CoV-2 spike protein.
The researchers used a weighted ensemble (WE) path sampling strategy to simulate atomistic pathways for the opening of the spike protein. WE focuses on the functional transitions between stable states rather than the states themselves. By running several parallel trajectories, the time spent in the stable state waiting for transitions over the energy barrier is minimized, making it more efficient than conventional MD simulations. The WE strategy generates continuous pathways providing the most direct, atomistic views for analyzing functional transitions and transient states that are too fast to be captured by laboratory experiments.
The team ran simulations on supercomputers in parallel for over a month, simulating 310 independent pathways. This included 204 pathways from the RBD down to up configuration and 106 pathways from the RBD down to open pathways. The open state included conformations that were similar to cryo-EM structures of the spike protein bound to ACE2.
The RBD in the down state is shielded by glycans at N165, N234, and N343, which have been reported to also stabilize the RBD in the up conformation. The WE simulations suggest the N343 glycan acts as a “glycan gate” and pushes up the RBD, and forms contacts with several residues of ACE2.
The authors initially saw this in several pathways by which spike protein opens and then analyzed all the 310 pathways where the N343 glycan formed contacts with the ACE2 residues to confirm this gating mechanism.
N343 glycan helps opening of RBD
To investigate the role of N343 glycan as a gating residue, the team used biolayer interferometry experiments, which evaluate the binding of the spike protein to ACE2 and how RBD opening is affected by mutations. Previous studies have suggested N165A and N234A mutations on the spike protein decrease ACE2 binding by 10% and 40%, respectively. They found a mutation in the N343 position, an N343A variant, decreased spike protein binding to ACE2 by almost 56%, more than those seen for the other variants.
This suggests the RBD up conformation is greatly affected by glycosylation at the N343 position. Further experiments identified R408A, D405A, and D427A as key spike protein residues that reduce ACE2 binding by about 13%, 27%, and 52%, respectively, suggesting these residues also help RBD opening.
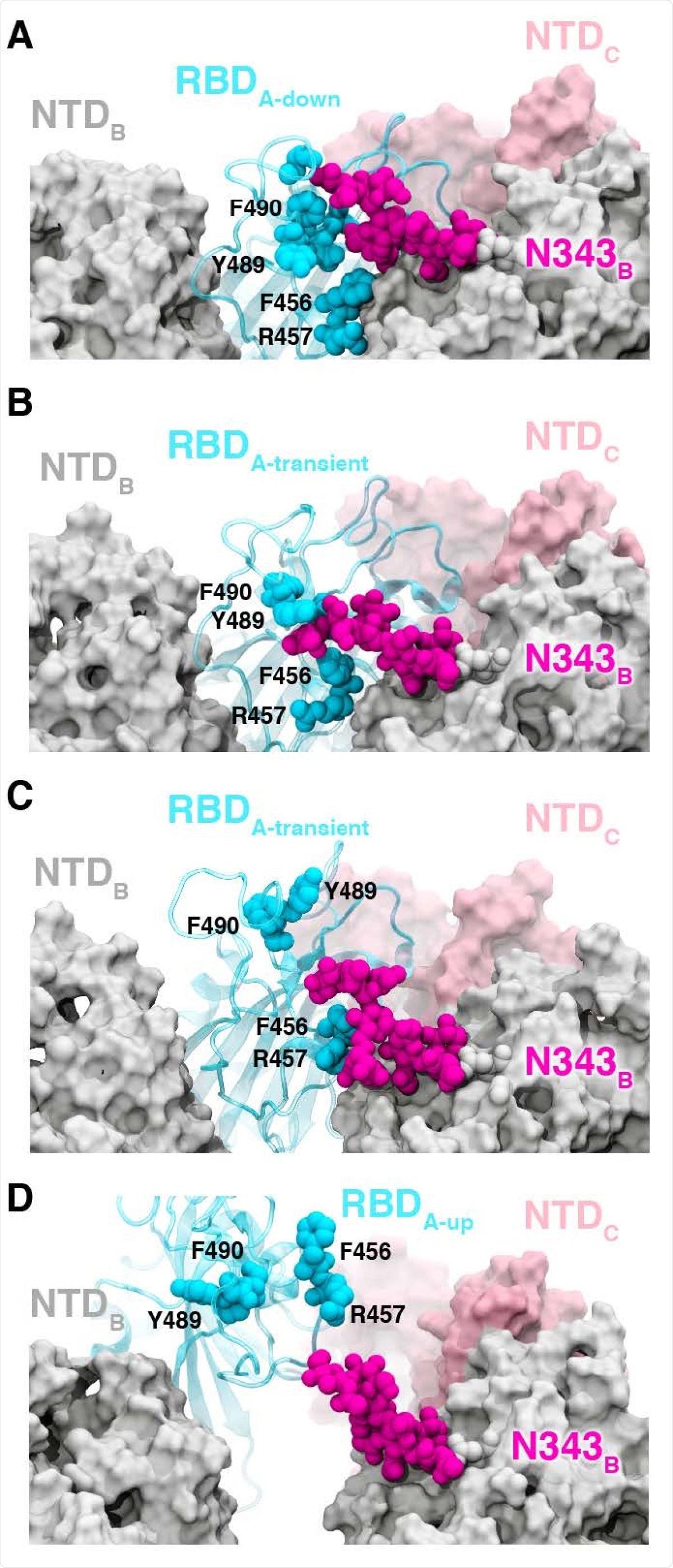
In the RBD down conformation, there is a hydrogen bond and several salt bridges between different residues. As the RBD opens, several salt bridges are broken and some are formed again with different residues until finally, there is only one contact between the RBD and the S1 subunit of the spike protein, the glycan at position N165.
According to the simulation results, the new SARS-CoV-2 variants, B.1, B1.1.7, B1.351, and P1, discovered recently do not have mutations in the residues that facilitate RBD opening. The simulation results indicate the glycan at N343 is critical in gating and aiding the RBD opening process.
*Important Notice
bioRxiv publishes preliminary scientific reports that are not peer-reviewed and, therefore, should not be regarded as conclusive, guide clinical practice/health-related behavior, or treated as established information.
- Sztain, T. E. et al. (2021) A glycan gate controls opening of the SARS-CoV-2 spike protein. bioRxiv. https://doi.org/10.1101/2021.02.15.431212, https://www.biorxiv.org/content/10.1101/2021.02.15.431212v1
Posted in: Medical Science News | Medical Research News | Disease/Infection News | Healthcare News
Tags: ACE2, AIDS, Angiotensin, Angiotensin-Converting Enzyme 2, Antibodies, Cell, Coronavirus, Coronavirus Disease COVID-19, Electron, Electron Microscopy, Enzyme, Glycan, Glycans, Glycosylation, Laboratory, Microscopy, Mutation, Protein, Receptor, Respiratory, Running, SARS, SARS-CoV-2, Severe Acute Respiratory, Severe Acute Respiratory Syndrome, Spike Protein, Syndrome, Virus

Written by
Lakshmi Supriya
Lakshmi Supriya got her BSc in Industrial Chemistry from IIT Kharagpur (India) and a Ph.D. in Polymer Science and Engineering from Virginia Tech (USA).
Source: Read Full Article
