Right now, in your body, lurk thousands of cells with DNA mistakes that could cause cancer. Yet only in rare instances do these DNA mistakes, called genetic mutations, lead to a full-blown cancer. Why?
The standard explanation is that it takes a certain number of genetic “hits” to a cell’s DNA to push a cell over the edge. But there are well-known cases in which the same set of mutations clearly causes cancer in one context, but not in another.
A good example is a mole. The cells making up a mole are genetically abnormal. Quite often, they contain a mutated DNA version of the BRAF gene that, when found in cells located outside of a mole, will often lead to melanoma. But the vast majority of moles will never turn cancerous. It’s a conundrum that has scientists looking to cellular context for clues to explain the difference.
Targeting the context, not just the mutations
The scientists say their results offer an important new perspective on cancer formation, one that contrasts with conventional wisdom.
“The standard idea that has been around for decades is that you basically need two types of DNA mutations to get cancer: an activated oncogene and a disabled tumor suppressor gene,” Dr. White says. “Once you clear those two hurdles, cancer will form. Now we have this entirely other thing—oncogenic competence—which adds a third layer to the mix.”
Dr. Baggiolini, a lead author on the study and a postdoctoral fellow in the Studer lab, likens the situation to starting a fire. “The DNA mutations are like a lit match: If you have the wrong wood or if the wood is wet, you might get a bit of flickering but no fire. But if you have the right wood, and maybe some kindling, the whole thing burns up.”
In this example, ATAD2 is the kindling. Developing a drug to remove this kindling would be another way to treat the cancer besides targeting the DNA mutations.
A technique with future potential
The hPSC techniques the team developed to study melanoma may prove to have wide applications for personalized cancer treatment. Already, Drs. White and Studer are using the technique to create disease models of individual patients’ cancer. From a patient’s blood, they can obtain cells to make hPSCs. They can then introduce to these cells the specific mutations that characterize the patients tumor. These genetically matched cells can then be used to test a large panel of drugs to see which may benefit the patient. These drugs can then be brought back to the zebrafish to ensure they really work in a living animal. The researchers believe this crossing over between cells in a dish and animal studies gives them the best chance of finding drugs that will work in patients.
“With hPSCs, we can potentially create patient-specific disease models of cancer for any different tissue in the body, be it from the brain or the liver or another organ,” Dr. Studer says. “I really hope that this will become a routine part of care.”
It’s likely no coincidence that these far-reaching results are the product of a steady, decade-long collaboration between two labs with different expertise. “It’s almost become a cliché to say that science is better in collaboration, but in this case it really was critical,” Dr. White says.
He adds: “Everyone wants science to go fast. We want science to go fast too. But sometimes science has to go slow to get to the core truth.”
To prove that ATAD2 was playing a decisive role, the scientists did additional experiments where they either took ATAD2 away or added it back in. When the scientists removed ATAD2 in a melanoma-prone zebrafish model, the cells lost their ability to form tumors. When they added it to MC cells, the cells gained this capacity. This told the researchers that ATAD2 was indeed a key lever of oncogenic competence.
Taking advantage of the vast amount of clinical data available at MSK and through the The Cancer Genome Atlas, they could show that ATAD2 is important in cancer: Patients with high amounts of ATAD2 have significantly worse survival, suggesting it plays an important role in determining the outcome of DNA mutations like BRAF.
“We’ve known for a while that cellular context matters in cancer formation,” says Richard White, a physician-scientist at Memorial Sloan Kettering Cancer Center who studies melanoma in the lab. “But exactly how context combines with genetic mutations to promote cancer is largely unknown.”
To get at that question, Dr. White teamed up with MSK developmental biologist Lorenz Studer, an expert at creating and using stem cells to study and treat disease. Through their complementary expertise—and the efforts of postdoctoral fellow Arianna Baggiolini and graduate student Scott Callahan—they were able to investigate how cancer genetics and developmental biology cooperate in cancer formation. A decade later, the results are in.
In a paper published September 3, 2021 in Science, Drs. White and Studer and their team report that melanoma formation depends on something called “oncogenic competence,” which is the result of a collaboration between the DNA mutations in a cell and the particular set of genes that are turned on in that cell. Cells that are competent to form melanoma are able to access a set of genes that normally are closed off to mature melanocytes (the cells that make melanin and give skin its color). In order to access these locked-up genes, the cells require specific proteins that act as keys. Without them, the cells do not form melanoma, even when they have cancer-associated DNA mutations.
The findings provide an explanation for why some cells, but not others, can form cancer, and offer potential therapeutic targets that could one day help patients.
From fish to humans
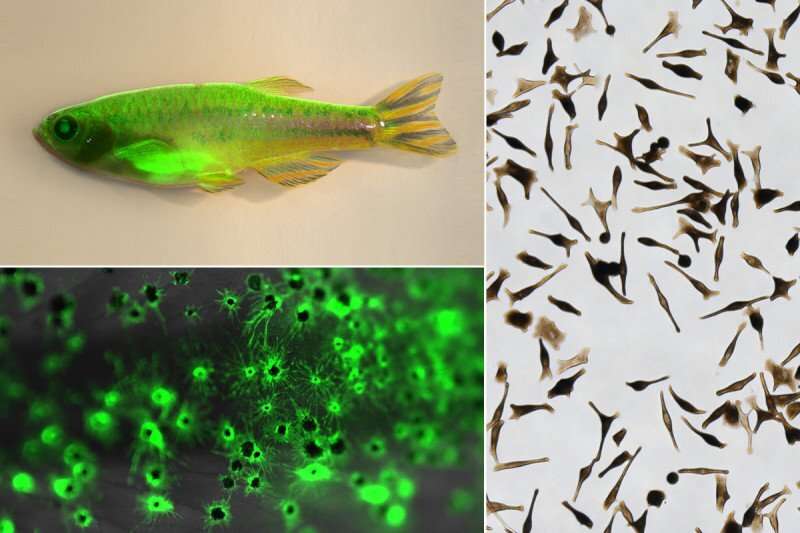
The collaborative project started more than a decade ago and began with an observation that Dr. White made while he was still a postdoc studying melanoma in zebrafish. These tiny fish are great for studying melanoma development because you can see tumors growing beneath the fish’s scales, and it’s easy to remove the tumors and study them at the molecular level.
“When we looked at these melanomas in zebrafish, we could see that there were a number of active genes that are more characteristic of embryonic cells rather than mature melanocytes,” Dr. White says. “We were curious why these genes were turned on. Are they important in the development of the melanoma, and if so how?”
To answer this question, Dr. White and his team genetically modified zebrafish to contain a mutated BRAF gene—the same one that is found in about one-half of melanomas. They introduced the BRAF gene in such a way that it would be turned on at three different stages of melanocyte development in different fish: at the neural crest stage (NC), the melanoblast stage (MB), and the melanocyte stage (MC). These stages refer to cell states that are progressively more differentiated. (You can think of the stages as akin to melanocyte kindergarten, elementary school, and high school.) Then, they let the fish grow up and watched for tumors.
After several months, they found that only the fish with BRAF activated at the NC and MB stages were capable of forming tumors (what the researchers call “oncogenic competence”). Cells with BRAF activated at the MC stage instead formed moles.
The result was striking. But what is true of fish isn’t necessarily true of humans. So, to extend these results, Dr. White teamed up with Dr. Studer to perform similar experiments on human cells. Dr. Studer’s team had previously shown that they could use human pluripotent stem cells (hPSCs) to make each of the three stages of melanocyte development. In this case, they introduced the mutated BRAF gene into hPSCs at the same three stages that were studied in the fish, and then implanted these cells in mice to see which were able to form tumors. Once again, only the first two stages—NC and MB—were consistently able to form tumors.
What controls ‘competence’?
Encouraged by these findings, the researchers went further to investigate a possible mechanism. Using what’s called “molecular profiling,” they compared what was different about the active genes in the three stages—for both the zebrafish tumors and the human stem cell-derived tumors. From this comparison, they were able to see that a key difference was a particular protein, ATAD2, that was active in NC and MB cells, but not in MC cells.
Source: Read Full Article
