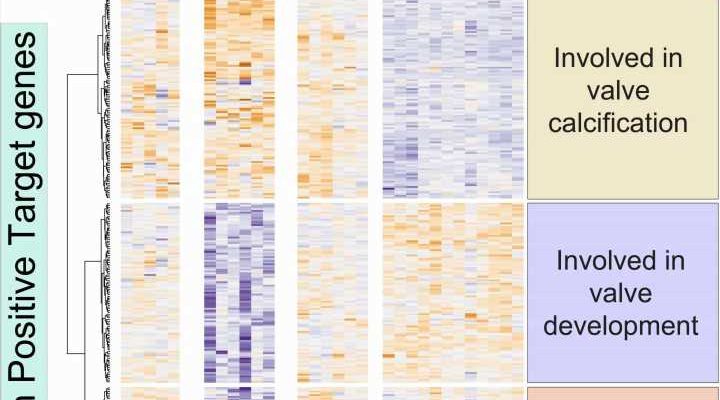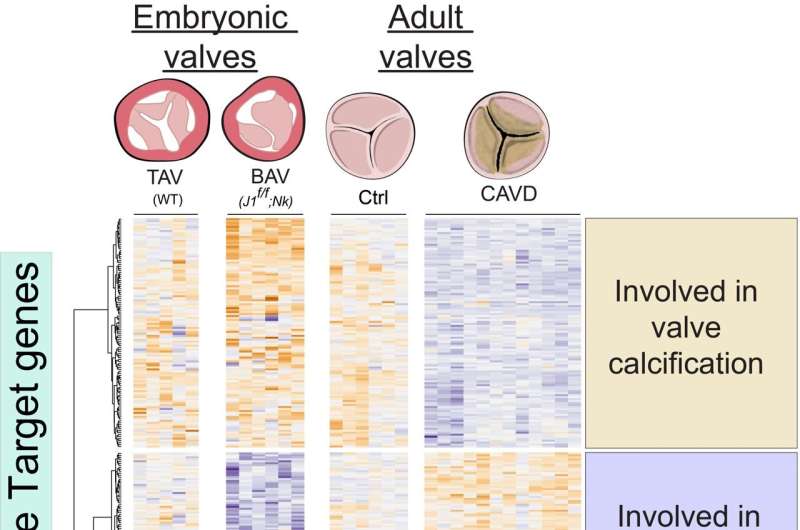
A team of researchers at the Centro Nacional de Investigaciones Cardiovasculares (CNIC) in Madrid has identified the molecular mechanisms that control the activity of genes involved in both the correct formation of the heart valves and the prevention of their subsequent calcification.
The findings, published in the journal Circulation Research, not only highlight the pathways through which the human heart forms but also offer clues for future medical advances. Study coordinator Dr. José Luis de la Pompa, who heads the Intercellular Signaling in Cardiovascular Development and Disease Laboratory at the CNIC, explained that “by understanding these fundamental processes, scientists are one step closer to solving the mysteries of the human heart and its pathologies.”
The heart is the body’s engine, and its correct function depends on the interaction of specialized components. One of these components is the endocardium, the single layer of cells that lines the inside of the heart, insulating the heart muscle from the blood it pumps throughout the body.
“But this is not its only function,” explained lead author and study co-coordinator Dr. Luis Luna Zurita, “This inner lining of the heart produces molecular signals that ensure the correct organization and function of the heart during embryonic development.”
For example, the endocardium plays an essential role in the formation of the heart valves.
“In response to different signals, cells in specific regions of the endocardium transform, acquire invasive properties, and colonize a territory beneath the myocardium, gradually forming structures called the valve primordia, which undergo progressive modeling to generate the mature adult heart valves,” said Dr. Luna Zurita.
Heart valves are crucial structures that regulate the one-way blood flow in the heart. One of the four heart valves is the aortic valve, located at the junction between the left ventricle and the aorta. In approximately 1% of the population, this valve has only two leaflets (bicuspid aortic valve, BAV) instead of the usual three (triscuspid aortic valve, TAV).
“This bicuspid valve is more prone to deteriorate and gives rise to a number of cardiovascular pathologies,” explained Dr. de la Pompa. One of these is valve calcification, which is frequent in these patients but also affects individuals with no apparent valve defect.
The accumulation of calcium deposits on the aortic valve affects its flexibility and function. One of the key consequences of calcification is valve stenosis, which restricts blood flow and compromises the heart’s pumping action, resulting in the need for surgery to replace the calcified valve.
They have established that many important steps in cardiovascular development are regulated by the Notch signaling pathway. Disruption of this highly conserved pathway causes both valve defects and valve calcification.
Notch acts on the endocardium at the earliest stages of development and during adult life. Mutations affecting different Notch pathway components are implicated in the formation of a BAV.
By carefully manipulating Notch pathway activity in isolated embryonic endocardial cells, Dr. de la Pompa’s team was able to identify the gene programs activated and inhibited by this pathway in the endocardium, distinguishing between an early and a late response.
To pursue their analysis further, the researchers needed to incorporate large databases from multiple sources. “Big data is becoming increasingly pivotal in numerous sectors, not least in biomedical research,” said Dr. Luna Zurita.
After creating a list of active genes in embryonic endocardial cells, Dr. Luna Zurita compared this with information on genetic programs in the embryonic and adult heart already described by Dr. de la Pompa’s lab. The comparison thus provided insight into valve formation during embryonic development and the calcification of adult valves, enabling the identification of key genes whose suppression disrupts normal valve formation in the embryo, is strongly associated with calcification in adulthood, or adversely impacts both processes.
The research also identified a set of regulatory regions that act on these genes. Dr. Luna Zurita explained that genes encoding proteins make up just 1% of the DNA in a mammalian genome. The regulatory regions are found in the remaining 99% and are responsible for a gene being expressed in a specific organ at a specific time.
“This implies that, while identifying changes in gene activity is obviously essential for understanding and treating disease, pinpointing the location of these genes’ regulatory regions is just as crucial. By analyzing changes in DNA compaction, we identified the regions that open or close in response to Notch pathway activity.”
Bioinformatics analysis and the integration of the databases from previous studies allowed the researchers to identify those DNA regions that likely control the expression of the genes implicated in heart valve formation and maintenance. In addition, this analysis provided crucial information about the molecules and signaling pathways responsible for this control.
One of the pathways identified was the Hippo signaling pathway, an extensively researched pathway that plays a crucial role in the control of cell division and organ size. “Our analysis revealed for the first time that cooperation between the Notch and Hippo pathways is required for the correct participation of the endocardium in valve formation,” concluded Dr. de la Pompa.
More information:
Luis Luna-Zurita et al, Cooperative Response to Endocardial Notch Reveals Interaction With Hippo Pathway, Circulation Research (2023). DOI: 10.1161/CIRCRESAHA.123.323474
Journal information:
Circulation Research
Source: Read Full Article