In a three-year study, researchers from Skoltech, Vavilov Institute of General Genetics of RAS, the Moscow-based Mental-Health Clinic No. 1 named after N.A. Alexeev and Serbsky Federal Medical Research Center of Psychiatry and Narcology have examined how gut microbes in patients with a major depressive disorder are different from those in mentally healthy people.
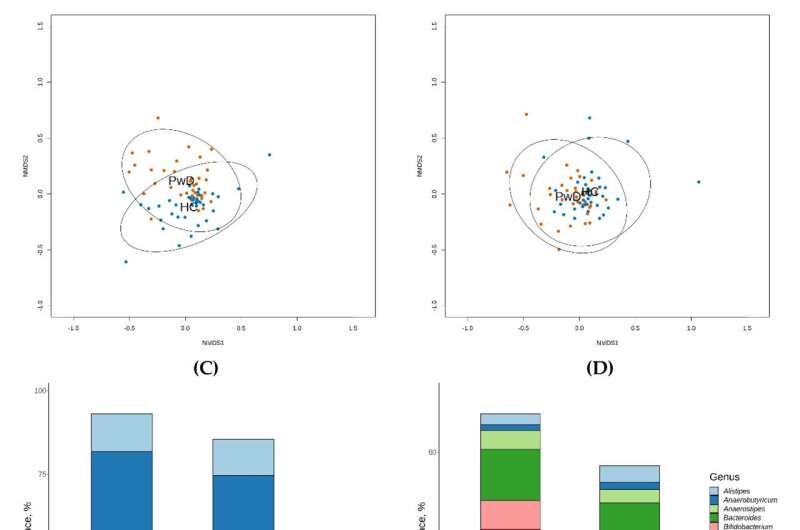
After analyzing all genes present in the gut microbiome, the team pinpointed a particular bacterium—Faecalibacterium prausnitzii—responsible for the largest functional discrepancy between the healthy and the depressed datasets. The findings, which hold promise for express mental health diagnostics and psychobiotic medications, are reported in Biomedicines.
“If you know which genes are represented to a lesser or greater extent in depressive patients compared with the healthy population, and which bacteria are responsible for this, you can attempt to do two things,” the study’s lead author, Skoltech Bio research intern Alexey Kovtun, commented. “First, you can use the analysis of fecal microbiota as a supplementary tool when diagnosing the mental disorder. Second, you can try to develop drugs that would ‘normalize’ gut microbiome in depressed patients.”
To identify “depressing” gut germs, the researchers conducted so-called full-metagenome analysis. That is, they recovered and sequenced the entirety of bacterial DNA from the feces samples of a cohort of patients diagnosed with the major depressive disorder and a cohort of mentally healthy individuals. “The result is that we know both which genes and bacterial species are present in the microbiome of each group, and how strongly they are represented,” Kovtun said.
The next step is identifying the set of genes that vary significantly between the healthy individuals and those with the mental disorder and zooming in on those particular genes to find out which of the bacteria carrying them are actually over- or underrepresented in the microbiome of depressed patients.
“One particular bacterium really stood out,” Kovtun explained. “It is called Faecalibacterium prausnitzii, and it is significantly less abundant in the gut of patients with the major depressive disorder. We linked it to three strongly underrepresented gene groups in the microbiome metagenome of those patients.”
The first of the three notable gene sets is involved in the production of the hormone melatonin, which regulates the sleep-wake cycle. The second one is associated with the formation of the classical neurotransmitters glutamate and gamma-aminobutyric acid. The third one is comprised by multiple genes responsible for the synthesis of short-chain acids, whose deficiency has been linked to depression.
According to the authors of the paper, the microbe whose role the study highlights—Faecalibacterium prausnitzii—has increasingly attracted the attention of researchers in the context of developing treatment options and diagnostic approaches for various diseases based on certain bacterial strains. The professional community is excited about this bacterium.
Source: Read Full Article
