Emergence of the Omicron variant of the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) (B.1.1.529) has startled the world. The new variant is the consequence of the rapidly mutating ability of SARS-CoV-2 which has already resulted in the generation of four variants of concern (VOC).
While the scientific community around the world is putting in their immediate efforts to ascertain the transmissibility, disease severity and susceptibility associated with Omicron, such endeavors demand the availability of Omicron virus isolate.
 Study: An in vitro and in vivo approach for the isolation of Omicron variant from human clinical specimens. Image Credit: Fit Ztudio/Shutterstock
Study: An in vitro and in vivo approach for the isolation of Omicron variant from human clinical specimens. Image Credit: Fit Ztudio/Shutterstock
Researchers from India have now successfully grown and isolated the Omicron variant from hamsters in vivo with further culturing in the Vero CCL-81 cells in vitro, in a step forward towards testing the effectiveness of existing SARS-CoV-2 vaccines and generation of new Omicron appropriate vaccines.
A pre-print version of the research paper is available on the bioRxiv* server while the article undergoes peer review.
Background
Omicron, the fifth VOC was first reported by South Africa in November 2021 followed by Botswana and Hong Kong. Since then, it has rapidly disseminated across the world.
India is currently witnessing a surge in coronavirus disease 19 (COVID-19) cases, which are believed to be driven by the Omicron variant. A total of 2,630 cases of the new variant surfaced in India by January 06, 2022.
Omicron, the heavily mutated SARS-CoV-2 variant, poses a serious public health concern due to the presence of mutations associated with enhanced binding affinity to ACE2, improved transmissibility, increased viral load, and escape from innate immunity.
Immediate studies are needed to evaluate the efficacy of already existing vaccines against the new variant in an effort to inform the public health authorities and healthcare providers. Therefore, the team led by Dr. Pragya Yadav undertook the current study in order to make the Omicron isolate available for the research community.
What did the researchers do?
Based on qPCR results, the oropharyngeal/nasopharyngeal swabs of SARS-CoV-2 positive international travelers (n=73) and contact case (n=1) were collected from Delhi and Mumbai airports in India during October 10, 2021 to December 13, 2021. Specimens were inoculated onto Vero CCL-81 cells.
The cultures were observed daily for any cytopathic effects (CPEs) and tissue culture infective dose 50 per cent (TCID50) values were calculated.
Two Omicron positive specimens [MCL-21-H-11828 (group-A) and MCL-21-H-12521/1 (group-B)] identified by whole-genome sequencing were selected for in vivo virus isolation.
Likewise, each specimen was inoculated into hamsters through the intranasal route. The nasal turbinate (NT) and lung specimens of passage 1 (P1) were further passaged into a new batch of hamsters (termed P2).
The NT and lung specimens, collected from hamsters on 3rd day-post-infection (DPI), were screened for the presence of virus using qPCR. Both P1 and P2 of SARS-CoV-2 positive NT and lung homogenized suspension were further inoculated onto Vero CCL-81 cells.
Next-generation sequencing was performed on clinical specimens, hamster specimens, and virus-infected Vero cells. Complete genome sequences of the Omicron variant, retrieved from clinical specimens, hamster specimens, and Vero cell cultures, were phylogenetically analyzed.
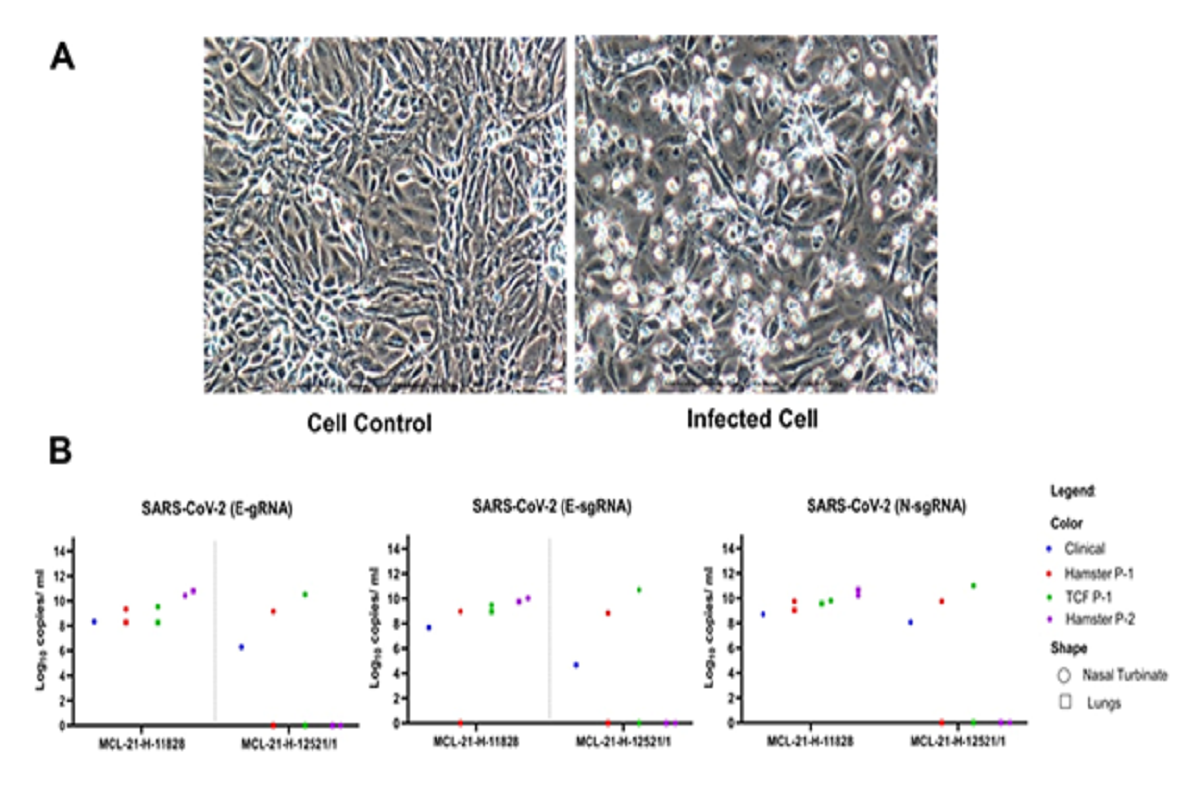 Figure 1: Omicron variant virus isolation in the Vero CCL-81 Cells and their viral titer: A) Vero CCL-81 cell control (Left) and Omicron infected Vero CCL-81 cell (Right) at passage 2, PID-4. B) The viral load of the SARS- CoV-2 in human clinical samples, hamster specimens at passage 1 and 2 and Vero CCL-81 at passage 1 culture. The genomic RNA and subgenomic RNA load for the E gene along with N gene from nasal turbinate and lungs of two different clinical specimens.
Figure 1: Omicron variant virus isolation in the Vero CCL-81 Cells and their viral titer: A) Vero CCL-81 cell control (Left) and Omicron infected Vero CCL-81 cell (Right) at passage 2, PID-4. B) The viral load of the SARS- CoV-2 in human clinical samples, hamster specimens at passage 1 and 2 and Vero CCL-81 at passage 1 culture. The genomic RNA and subgenomic RNA load for the E gene along with N gene from nasal turbinate and lungs of two different clinical specimens.
What did the researchers find?
Although, a previous study from Hong Kong reported the isolation of the Omicron variant in vitro, this is the first study reporting the isolation using an initial in vivo and subsequent in vitro approach.
Of all the clinical samples collected in a two-month span, fifteen were identified as Omicron, of which thirteen were asymptomatic cases, while two had a mild cold, cough, and body ache.
Initial efforts to culture the virus, from Omicron positive clinical specimens, on Vero CCL-81 cells failed. It included three blind passages for each sample, however, neither the virus could be detected in qPCR nor any CPEs were observed on Vero cells.
However, in vivo isolation from clinical specimens demonstrated active viral replication in both NT and lung specimens of group-A hamsters, while only NT showed virus replication in hamsters from the group-B. The NT and lungs of hamsters group-A demonstrated 2.0–2.2×109 and 2.3×105–1.9×108 genomic viral RNA (gRNA) copies per ml respectively, while the NT of hamsters from the group-B had 1.5×108–8.3×109 gRNA copies. Further, the P2 passaging of NT and lung specimens of group-A hamsters demonstrated 2.6×1010 and 6.6×1010 gRNA copies per ml in NT and lungs respectively on 3rd DPI.
Although the direct in vitro culturing, on Vero CCl-81 cells, from clinical samples failed, but when homogenized hamster NT and lungs specimens of group-A were used for culture inoculation, typical CPE in P-1 and P-2 of Vero CCL-81 were observed with 3.4×109 to 1.9×1010 and 1.8×108 to 1.0×109 gRNA copies per ml respectively. Only hamster NT specimen of group-B displayed CPEs in Vero CCL-81 with 3.3×1010 and 4.8×1010 gRNA copies.
Single nucleotide variations (SNV) were observed in the Omicron sequences from the human clinical, hamster and Vero CCL-81 samples. Insertion of AGT nucleotides at genomic position 22,195 was observed in the hamster P1 sequence and this insertion resulted in alteration of two amino acid encoding codons. The tyrosine at 69 amino acid position of spike was lost in the P1 hamster and observed in the P2 hamster.
Substitution mutation N440K, that has been reported to be associated with higher infection titers as well as neutralization escape, was observed with varying frequencies in clinical, hamster and Vero virus culture samples. One hundred percent of the hamster amino acid sequences and 68.5% of the human clinical sample amino acid sequences demonstrated this mutation. The Vero CCL-81 virus culture derived from P1 of the hamster demonstrated the substitution in 86.0% of the sequences, while Vero CCL-81 virus culture derived from of the P2 of the hamster demonstrated substitution in 71.79% sequences.
Besides these SNVs, the viral sequences from human clinical specimens, hamster specimens and Vero CCL-81 passages didn’t show any mutations in the Furin cleavage site during sequential passages. Based on the observation, the team suggests that such virus isolates would maintain their pathogenicity similar to the original specimen.
“The isolated Omicron virus would be useful in new vaccine development, vaccine efficacy studies, pathogenicity in hamsters, antiviral research and estimation of virus-neutralizing titers in vaccinated and recovered individuals”, the team concludes.
 Figure 2: SNV and phylogenetic tree of the Omicron clinical and its isolate sequences: A) Single nucleotide variation in the S gene and its frequency in different in vivo and in vitro sequences. The x axis is the SNVs and Y axis marks the frequency. B) Neighbour joining tree with a bootstrap replication of 1000 cycles to assess statistical robustness. Different SARS-CoV-2 variants are marked in various colors.
Figure 2: SNV and phylogenetic tree of the Omicron clinical and its isolate sequences: A) Single nucleotide variation in the S gene and its frequency in different in vivo and in vitro sequences. The x axis is the SNVs and Y axis marks the frequency. B) Neighbour joining tree with a bootstrap replication of 1000 cycles to assess statistical robustness. Different SARS-CoV-2 variants are marked in various colors.
*Important notice
bioRxiv publishes preliminary scientific reports that are not peer-reviewed and, therefore, should not be regarded as conclusive, guide clinical practice/health-related behavior, or treated as established information.
- Yadav P.D., et al. (2022). An in vitro and in vivo approach for the isolation of Omicron variant from human clinical specimens. bioRxiv. doi: https://doi.org/10.1101/2022.01.02.474750 https://www.biorxiv.org/content/10.1101/2022.01.02.474750v1
Posted in: Medical Science News | Medical Research News | Disease/Infection News
Tags: ACE2, Amino Acid, binding affinity, Cell, Cold, Coronavirus, Coronavirus Disease COVID-19, Cough, covid-19, Efficacy, Genome, Genomic, Healthcare, immunity, in vitro, in vivo, Lungs, Mutation, Nasopharyngeal, Nucleotide, Nucleotides, Omicron, Public Health, Research, Respiratory, RNA, SARS, SARS-CoV-2, Syndrome, Tissue Culture, Tyrosine, Vaccine, Virus

Written by
Namita Mitra
After earning a bachelor’s degree in Veterinary Sciences and Animal Health (BVSc) in 2013, Namita went on to pursue a Master of Veterinary Microbiology from GADVASU, India. Her Master’s research on the molecular and histopathological diagnosis of avian oncogenic viruses in poultry brought her two national awards. In 2013, she was conferred a doctoral degree in Animal Biotechnology that concluded with her research findings on expression profiling of apoptosis-associated genes in canine mammary tumors. Right after her graduation, Namita worked as Assistant Professor of Animal Biotechnology and taught the courses of Animal Cell Culture, Animal Genetic Engineering, and Molecular Immunology.
Source: Read Full Article
