Testing a population for multiple genes at once will be limited in its ability to accurately predict disease risk, argues a new paper involving UCL’s Professor Aroon Hingorani.
DNA sequence commonly varies between individuals at many different points throughout the genome. Some of these genetic variants influence individual risk of common diseases such as dementia, cancer, diabetes, and cardiovascular disease, through effects on the expression or function of the encoded proteins. The effects of these variants on disease risk in any individual can be summarized by generating a polygenic risk score.
Strong claims have been made about the transformative potential of polygenic risk scores for prediction, prevention, and early detection of disease and such testing has attracted the interest of policy makers and the commercial sector.
However, in an analysis article in the British Medical Journal, clinical researchers from UCL, The Institute of Cancer Research, London, and the University of Oxford argue that the predictive performance of such tests is limited.
Co-author Professor Aroon Hingorani (UCL Institute of Cardiovascular Science), who is also Cardiovascular Theme Lead for the UCLH NIHR Biomedical Research Center and a Consultant in Internal Medicine at UCLH, said, “Population-wide ‘polygenic scoring’ is inherently limited because many cases of disease occur among people who do not have high polygenic scores and many of those with high polygenic scores do not develop disease.”
NHS genetic testing currently focuses on a small number of well-understood disease mutations which strongly raise the risk of disease—such as BRCA1 and BRCA2 in breast and ovarian cancer.
Polygenic scoring is a different type of genetic test which instead looks across thousands of common variants in a person’s DNA, each of which will individually have only a small impact on risk, to gain a collective assessment of the genetic risk of disease.
Two recent Government reports show marked enthusiasm for polygenic scores in health care and assert that they offer a “step change”‘ in screening for disease.
The NHS is now partnering with the UK’s largest research program, Our Future Health, to offer risk information based on polygenic scores to five million people in the UK population. Such information is expected to inform clinical decision-making including access to screening.
Polygenic scores should be ‘carefully evaluated’ in large studies
The authors argue that the risks and benefits of genetic testing must be carefully evaluated in large studies. Their analysis concludes that initiatives which use polygenic scores to screen for people at risk of common illnesses such as cancer and heart disease will miss most cases in the population.
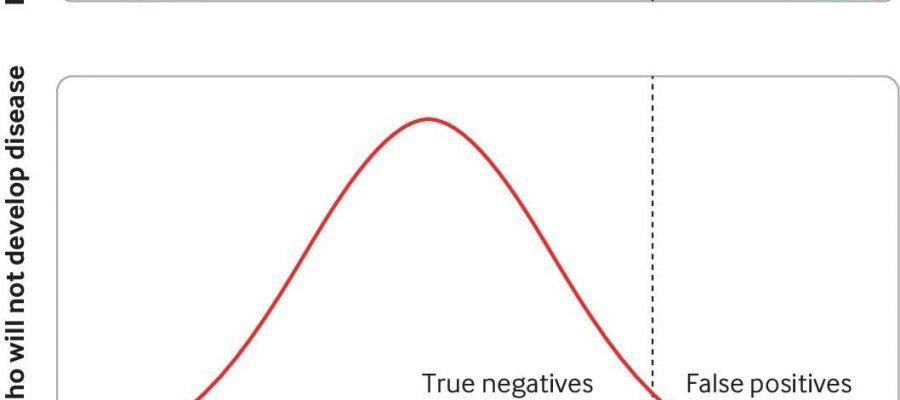
When screening the population for a disease using polygenic scores, many have advocated for using a high polygenic score to target screening or preventative strategies for disease. According to the researchers doing so would miss most of the disease in the population and result in a sizeable number of healthy individuals undergoing invasive tests and therapies who will never go on to develop the disease.
For example, NICE uses a 17% lifetime risk of breast cancer as the threshold for deeming women at “moderate risk.” But in a polygenic risk score study which also used this threshold, only 39% of women who will go on to develop breast cancer have moderate or high-risk scores, meaning the majority of breast cancer cases are missed using these scores. Meanwhile, 22% of women who will not develop breast cancer will have a high polygenic risk score and will therefore be a “false positive.”
Putting scores in context
The scientists argue that since polygenic risk scores only indicate a small proportion of overall disease risk, that they should be presented in context, with the normal background disease risk communicated for comparison to patients and clinicians.
For example, women in the top 5% of polygenic scores for breast cancer have a 19% lifetime risk of developing the disease, compared with a population risk of 11.8%.
For prostate cancer, those with a polygenic risk score in the top 5% carry a 22% risk compared with a 12.7% population risk.
The differences become even more modest in less common cancers. For example, people in the top five% of polygenic scores for ovarian cancer have a lifetime risk of 2.1%, compared with a population risk of 1.6%.
The authors outline various additional concerns with widespread use of polygenic scores to inform clinical decision making—including the cost and complexity of screening and diagnostic pathways, and the risk that they will expose large numbers of people with “high” polygenic scores to often unnecessary follow-up tests.
Lead author Dr. Amit Sud, Academic Clinical Lecturer in Genetics and Epidemiology at The Institute of Cancer Research, London, said, “There is a huge amount of enthusiasm about polygenic scores, and they do have the potential to improve our ability to predict who will or will not develop a disease, albeit rather modestly.
“But we argue that the benefits and harms surrounding the use of polygenic scores are carefully evaluated before they are widely implemented. Given most of the disease in a population occurs in people who are not at high polygenic risk, these scores should not detract from effective population-wide screening and interventions to address modifiable and impactful risk factors like smoking and socioeconomic deprivation.”
More information:
Amit Sud et al, Realistic expectations are key to realising the benefits of polygenic scores, BMJ (2023). DOI: 10.1136/bmj-2022-073149
Journal information:
British Medical Journal (BMJ)
Source: Read Full Article