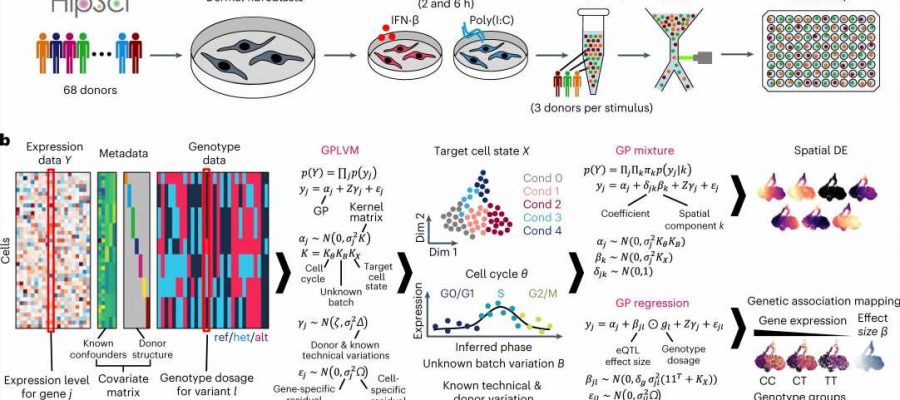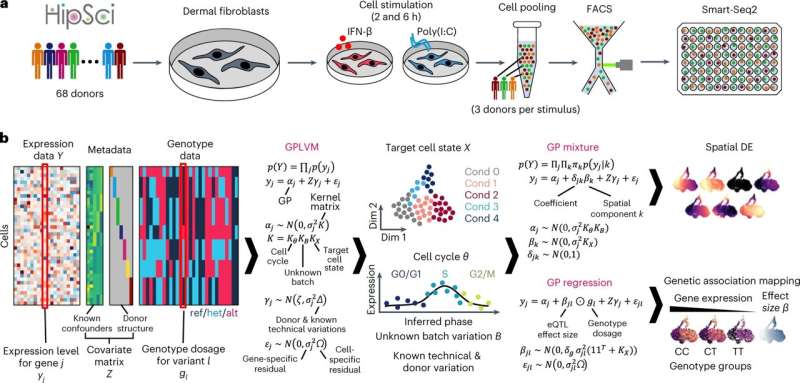
Researchers have discovered a mechanism for COVID-19 susceptibility using a newly created tool. The tool, GASPACHO, captures dynamic changes in gene expression along the innate immune response, allowing researchers to identify genes and molecular pathways associated with disease risk that have previously been too complex to detect or interpret.
Using GASPACHO (GAuSsian Processes for Association mapping leveraging Cell HeterOgeneity), researchers at the Wellcome Sanger Institute, the National Center for Child Health and Development in Japan, Tel Aviv University and their collaborators have identified a gene variant that affects COVID-19 susceptibility.
Understanding genetic factors contributing to COVID-19 infection and severity may provide new biological insights into disease pathogenesis and identify therapeutic targets. It is hoped the tool can be applied to discover further susceptibility mechanisms across other human disorders.
The study, published in Nature Genetics, helps to unpick the relationship between specific genes, their expression levels, and their potential connection in disease susceptibility. The team highlight the utility of the tool with a COVID-19 case study.
There exists a wide variation in how people respond to COVID-19. Around 80 percent of infected people experience a mild-to-moderate bout of illness, while some will experience mainly respiratory symptoms that are much more severe, requiring hospitalization and even intensive care. Part of this variation may be down to differences in our genes, specifically differences in our genetic regulation of gene expression.
The regions that affect gene expression are called expression quantitative trait loci (eQTLs). These are like signposts in our DNA that indicate which genetic variations are linked to changes in the expression of certain genes, affecting how much or how little a gene is dialed up or down, leading to differences in the levels of proteins produced by that gene.
While genome-wide association studies (GWAS) have identified numerous disease-associated variants involved in gene expression, implicating the involvement of eQTLs, they are unable to show any causal relationships. Genome-wide eQTL mapping however, has shown potential in revealing underlying genetic mechanisms of variation in disease outcomes.
In the new study, scientists set out to explore patient-specific immune responses through mapping eQTLs. They employed a novel approach to show how genetic variation within cells affects the overall immune response across individuals.
Researchers from the Wellcome Sanger Institute and their collaborators in Japan and at Tel Aviv University triggered an antiviral response in human fibroblast cells from 68 healthy donors, then profiled them using single-cell transcriptomics to put GASPACHO to the test.
The tool uses non-linear regression modeling to capture dynamic changes in eQTLs occurring at different stages of the immune response. Unlike previous eQTL mapping attempts that aggregate single-cell data—measuring average gene expression over many cells—GASPACHO enables cell-specific resolution to track changes over time and across individual cells.
The team identified 1,275 eQTLs within the genome which alter gene expression along the innate immune response between people, relevant for 40 immune-related diseases such as Crohn’s disease and diabetes.
The researchers found that when applying the tool to investigate variation in COVID-19 outcomes, lower expression of OAS1 gene variation occurred in those more likely to get COVID-19. The OAS1 gene encodes a protein involved in clearing viral RNA from the cell.
In COVID-19 patients, the team found lower OAS1 expression in nasal epithelial cells as well as monocytes in blood—both viral target cell types—compared to a reference genotype group. Their findings suggest that OAS1 expression can be modulated by a common splicing variant, OAS1 splicing QTL, at these target cell types. This is a genetic alteration in the DNA sequence at the boundary of an exon and intron. In these cells, the splicing variant will likely directly influence the efficacy of viral RNA clearance in the individual, explaining the impaired clinical outcome in the COVID-19 patient group.
While this genetic alteration needs to be explored further to fully understand the role it plays, it offers insights into the molecular mechanisms underlying susceptibility to COVID-19 and other immune-related diseases, providing a basis for developing potential therapies harnessing these genetic mechanisms.
Dr. Natsuhiko Kumasaka, first author of the study from the National Center for Child Health and Development in Japan, said, “We may in the future be able to use OAS1 and other genes on the same cascade in drug discovery or as therapeutic targets, but more research is needed to understand the specific mechanisms by which OAS1 or related genes may contribute to COVID-19.”
Dr. Tzachi Hagai, co-lead author of the study from Tel Aviv University, said, “It’s remarkable how small differences in our genetic makeup can affect our health and susceptibility to disease, just by influencing how active our genes are. While host-specific genetic factors are only one part of the puzzle, our work sheds light on the molecular mechanisms underlying various traits, diseases, and drug responses and how these may interact among wider environmental, clinical and social factors.”
“The findings here underscore the importance of ongoing scientific investigations to unravel the complex interactions between human genetics and the outcome of pathogen infection, including by emerging viruses such as SARS-CoV-2.”
Dr. Sarah Teichmann, co-lead author of the study from the Wellcome Sanger Institute and co-chair of the Human Cell Atlas organizing committee, said, “This new tool will be important in extracting meaningful insights from the huge amount of data that the Human Cell Atlas is generating, in its aim to map every cell type in the human body. Using the tool, we hope to uncover many underlying genetic mechanisms, and ultimately drug targets to aid in the development of new treatments for a variety of diseases.”
More information:
Natsuhiko Kumasaka et al, Mapping interindividual dynamics of innate immune response at single-cell resolution, Nature Genetics (2023). DOI: 10.1038/s41588-023-01421-y
Journal information:
Nature Genetics
Source: Read Full Article