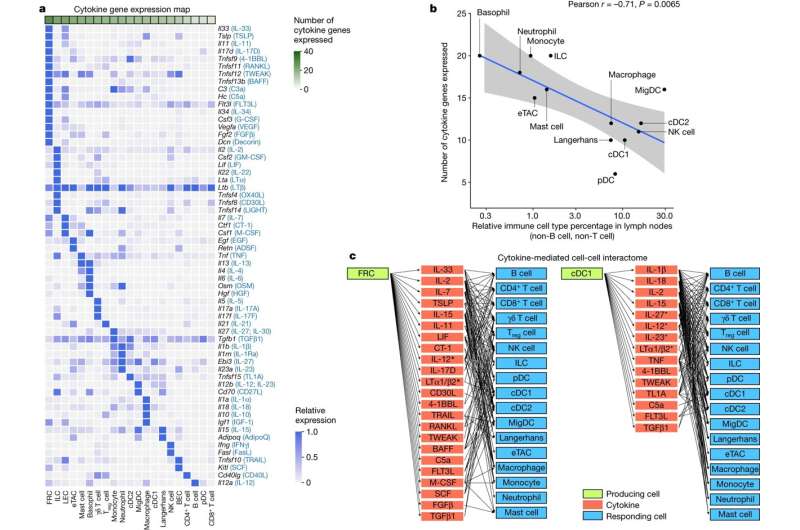
The immune system can carry out many biological processes, from killing viruses to fighting cancer, thanks in large part to approximately 100 key cell-signaling proteins called cytokines, which instruct immune cells what to do. Cytokines are also targeted by drugs for many diseases such as rheumatoid arthritis, COVID-19, and cancer, but until now, scientists haven’t had a comprehensive view of how different immune cells respond to different cytokines because the immune system is so complex.
A new, large-scale reference created by researchers at the Broad Institute of MIT and Harvard could help scientists and clinicians better understand the role of cytokines in health and disease. The reference, called the Immune Dictionary, appears in Nature.
Using single-cell RNA sequencing to analyze gene expression in individual cells, the researchers have found how 86 major cytokines affect 17 immune cell types in mice. They found a surprising level of complexity in the immune system: Cytokines can trigger more immune responses, and immune cells can perform more functions than previously thought.
The team also developed software for scientists, called Immune Response Enrichment Analysis (IREA), that they can use to identify the most active cytokines involved in a disease or drug response, and how different immune cells carry out different functions depending on which cytokine signal they receive.
We spoke with Ang Cui, first and co-corresponding author on the study, formerly a postdoctoral researcher in Nir Hacohen’s lab at Broad and now an assistant professor at the Harvard School of Dental Medicine and Harvard Medical School (HMS); and Hacohen, a co-corresponding author, institute member at the Broad, and professor of medicine at HMS and Massachusetts General Hospital; about what makes their immune dictionary unique and how it could impact immunology research in this Q&A.
What makes this study significant?
Nir Hacohen: This is the first single-cell resolution dictionary of each major immune cell type responding to each major cytokine in vivo at an unprecedented scale.
Typical studies of immune responses may look at roughly five immune cell types in a couple of conditions, whereas this study looked at nearly all major immune cell types responding to nearly all of the major cytokines. That’s more than 1,400 cytokine-cell type combinations—two orders of magnitude larger than typical studies, allowing the team to comprehensively document the complexity of the immune system.
The scale was unique, but also, doing this at single-cell resolution was critical because it turns out that if you isolate a macrophage, there are actually different types of monocytes and macrophages.
Single-cell resolution allows for a more natural view of those subsets. We also made the decision to do this in the mouse rather than in culture. There have been a lot of papers over the years where people take cells and stimulate them with cytokines in dishes in the lab, but we never knew what it meant in an animal. This was the first time we’ve done it in a physiological context.
Ang Cui: This research could have far-reaching implications, accelerating our understanding of natural immune responses and human immune-related diseases. For many immune-mediated diseases, there’s no cure or treatment. Some patients develop resistance to treatment and we cannot predict who they will be.
There are many open questions, and having this foundational reference can significantly improve our understanding of the immune system. In the future, when doctors give patients cytokine therapies targeting the immune system, they could potentially look in the Immune Dictionary for expected cell-level immune responses.
Did any results surprise you?
Ang Cui: We found that both the immune responses to cytokines and the plasticity of immune cells were much more complex than we previously appreciated. Even IL-1β, one of the first discovered and most well-studied cytokines, induces much more complex responses than previously known. IL-1β can induce distinct responses in each cell type, which shows how a single cytokine can trigger a coordinated, multicellular immune response. This could only have been revealed by creating this kind of global map of every immune cell response to the cytokine.
Nir Hacohen: The dictionary gave us many surprises. I was shocked by what we found about IL-1β. I think for many people, this will help them make sense of this central cytokine.
Ang Cui: The study also showed us how plastic immune cells are. For example, macrophages are well known to be polarized into M1-like (proinflammatory) or M2-like (reparative) states (which helps determine their function), but we didn’t know how other immune cell types are polarized. We found that every immune cell type can be polarized into diverse states depending on which cytokines they receive. Natural killer cells, for example, can perform different functions depending on which cytokines we give them.
How do you hope other scientists will use these findings?
Ang Cui: Long-term, a more precise understanding of in vivo immune responses could enable precision medicine, overcoming current limitations for creating effective immunotherapies for a wide range of diseases. The dictionary could also be used for basic biology studies, if you want to know what cytokines can trigger a specific gene, for example.
We’ve already used our software to interrogate the immune response in four diseases: lupus, cancer, COVID-19, and hepatitis C. It identified the most active cytokines for each disease.
Nir Hacohen: If I were an immunologist working on my single-cell dataset in any disease or in any process, I would want to run a cytokine analysis using this new reference, because cytokines drive so many of the decisions.
I hope our approach helps scientists make sense of any immune process, vaccine, disease, or therapy response, so that one can infer which cytokines are contributing to it. You could then block those cytokines in animal models and see what their roles are. We’re excited to see what people will do with it.
More information:
Ang Cui et al, Dictionary of immune responses to cytokines at single-cell resolution, Nature (2023). DOI: 10.1038/s41586-023-06816-9
Journal information:
Nature
Source: Read Full Article